Gene
KWMTBOMO11583 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004023
Annotation
Cyclin_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.587
Sequence
CDS
ATGAAAGACTTAGTTGACGTAATGGCTTTGCAAAACAGACGTGAAAAGCGCGTACCCGACTATCGAGGCGCGCCGAACCACAGCTTAGCAACAAATTTTGTTTTTGAATGTGGAATAAAGCTTGGCTTGCAGCCTGCAACTATAGCAACTGCTGCCATATTTTACCACAAATTCTTCAAAGAGGCCGACAAGAATGATTACGACTGCTACGTAATCTGCACTGCTTGTTTGTGTGCAGCCGGGAAATCTCGAGATGAGCCTGTTAGGCTGCGAGATGCTGTAAATGTAGCACACAATTCGATAAATCGTGGAGCCGGTCCGTTGGAGTTAGGCGACGAGTACTGGTCATGGCGAAGTGCTGTAGCTCAAGCCGAGCTTCTTGTGCTGAGACTGCTGGGATTTAATCTGGAAGCGCCGTCGCCACACCGGTATCTATTGCATTATCTCAAATCGCTGCAAGAATGGTTTCCAGTAGCGCAGTGGCGTTCAGCTCCCATTGCTCGAATGGCAATGGCTTTCCTTCAGGATTTCCATCACTCACCAGCAGTACTTGATTATCGAGCACCCCATATAGCAGTGGCGTGTTTAACTTTAGCTTTACATGTATTAGGAGTATCAGTGCCATTGGCATCCACACTAGATGATGATGCTGCATGGTGTTCAGTTTTCACAAAGGATCTGCAGAAAGAAAAAAATTGGGAAATAATGGAGAAGATAATGCAAGTTTATGGTCGTGAACCGGAACCTTTGTAA
Protein
MKDLVDVMALQNRREKRVPDYRGAPNHSLATNFVFECGIKLGLQPATIATAAIFYHKFFKEADKNDYDCYVICTACLCAAGKSRDEPVRLRDAVNVAHNSINRGAGPLELGDEYWSWRSAVAQAELLVLRLLGFNLEAPSPHRYLLHYLKSLQEWFPVAQWRSAPIARMAMAFLQDFHHSPAVLDYRAPHIAVACLTLALHVLGVSVPLASTLDDDAAWCSVFTKDLQKEKNWEIMEKIMQVYGREPEPL
Summary
Similarity
Belongs to the cyclin family.
Uniprot
H9J3D6
A0A3S2NSW2
A0A0L7LUU8
A0A1E1WJR3
A0A2A4JFT3
S4PIP3
+ More
A0A194QIM7 A0A194QXD2 A0A2H1X382 A0A212EIF2 D6WSE2 A0A2J7R5I9 A0A2J7R5I2 A0A067QJ65 A0A1W4X2Z6 A0A1Y1NI35 V5GLU9 J3JUG7 U5ESQ3 A0A0L0C0X5 A0A182GP49 B0WAB9 Q17K96 A0A1Q3EWT2 A0A336M3T8 A0A1B0A2D7 D3TM20 A0A1A9Y0Y7 A0A1B0B6X7 A0A1A9WD88 A0A1I8Q3R2 A0A1A9VFA3 A0A0K8TSI0 W8BMY0 A0A1B6EBY4 A0A1B6F6L5 A0A1I8MAC3 A0A0J7NL68 A0A1B6LS32 F4X6W6 A0A195B247 E0VQY8 A0A158NFX6 A0A2A3E4S4 E1ZYZ9 A0A088A6G8 V9IKZ2 E9J652 A0A0L7R8K7 E2BX29 A0A026WPQ2 A0A195E6R4 A0A151XC35 A0A154PJA8 A0A1A9X4T7 A0A0C9RMH0 A0A0M8ZPM1 A0A195F1J6 A0A1B6IKK8 A0A1B0CLM9 A0A1B6IYB4 A0A182KA64 A0A182V0C5 A0A0K8TXS4 A0A1L8DWT6 A0A084WCT5 A0A1L8DWS9 A0A182IVR9 A0A182LB61 Q7Q9T2 A0A182HPD5 A0A182TRE1 K7ILS7 A0A182X0W7 A0A182M4V5 B4J8Q0 Q7QLK2 A0A182WMB6 A0A182STZ8 A0A1A9V274 A0A182YJ16 A0A0Q9W541 B3LW96 B4KRI4 A0A1W4U8C8 A0A023EK93 B4PLX5 A0A182NZQ7 A0A1B0AF92 A0A182NLX0 A0A3B0KQ92 Q299M1 Q9VE72 Q7YU34 A0A182Q8V8 Q8IG98 B4NH39 B4LQ75 A0A2P8YYL4 A0A146KQP8
A0A194QIM7 A0A194QXD2 A0A2H1X382 A0A212EIF2 D6WSE2 A0A2J7R5I9 A0A2J7R5I2 A0A067QJ65 A0A1W4X2Z6 A0A1Y1NI35 V5GLU9 J3JUG7 U5ESQ3 A0A0L0C0X5 A0A182GP49 B0WAB9 Q17K96 A0A1Q3EWT2 A0A336M3T8 A0A1B0A2D7 D3TM20 A0A1A9Y0Y7 A0A1B0B6X7 A0A1A9WD88 A0A1I8Q3R2 A0A1A9VFA3 A0A0K8TSI0 W8BMY0 A0A1B6EBY4 A0A1B6F6L5 A0A1I8MAC3 A0A0J7NL68 A0A1B6LS32 F4X6W6 A0A195B247 E0VQY8 A0A158NFX6 A0A2A3E4S4 E1ZYZ9 A0A088A6G8 V9IKZ2 E9J652 A0A0L7R8K7 E2BX29 A0A026WPQ2 A0A195E6R4 A0A151XC35 A0A154PJA8 A0A1A9X4T7 A0A0C9RMH0 A0A0M8ZPM1 A0A195F1J6 A0A1B6IKK8 A0A1B0CLM9 A0A1B6IYB4 A0A182KA64 A0A182V0C5 A0A0K8TXS4 A0A1L8DWT6 A0A084WCT5 A0A1L8DWS9 A0A182IVR9 A0A182LB61 Q7Q9T2 A0A182HPD5 A0A182TRE1 K7ILS7 A0A182X0W7 A0A182M4V5 B4J8Q0 Q7QLK2 A0A182WMB6 A0A182STZ8 A0A1A9V274 A0A182YJ16 A0A0Q9W541 B3LW96 B4KRI4 A0A1W4U8C8 A0A023EK93 B4PLX5 A0A182NZQ7 A0A1B0AF92 A0A182NLX0 A0A3B0KQ92 Q299M1 Q9VE72 Q7YU34 A0A182Q8V8 Q8IG98 B4NH39 B4LQ75 A0A2P8YYL4 A0A146KQP8
Pubmed
19121390
26227816
23622113
26354079
22118469
18362917
+ More
19820115 24845553 28004739 22516182 23537049 26108605 26483478 17510324 20353571 26369729 24495485 25315136 21719571 20566863 21347285 20798317 21282665 24508170 30249741 24438588 20966253 12364791 14747013 17210077 20075255 17994087 9087549 25244985 24945155 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 26823975
19820115 24845553 28004739 22516182 23537049 26108605 26483478 17510324 20353571 26369729 24495485 25315136 21719571 20566863 21347285 20798317 21282665 24508170 30249741 24438588 20966253 12364791 14747013 17210077 20075255 17994087 9087549 25244985 24945155 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 26823975
EMBL
BABH01021563
RSAL01000096
RVE47711.1
JTDY01000042
KOB79215.1
GDQN01003811
+ More
GDQN01002510 JAT87243.1 JAT88544.1 NWSH01001566 PCG70840.1 PCG70841.1 GAIX01000124 JAA92436.1 KQ459185 KPJ03306.1 KQ461073 KPJ09615.1 ODYU01013106 SOQ59791.1 AGBW02014665 OWR41255.1 KQ971352 EFA06631.1 NEVH01006994 PNF36090.1 PNF36088.1 KK853284 KDR08981.1 GEZM01004710 JAV96325.1 GALX01005889 JAB62577.1 APGK01028620 BT126880 KB740694 KB632300 AEE61842.1 ENN79461.1 ERL91750.1 GANO01003154 JAB56717.1 JRES01001049 KNC25973.1 JXUM01077808 KQ563028 KXJ74617.1 DS231870 EDS41009.1 CH477227 EAT47089.1 GFDL01015268 JAV19777.1 UFQT01000523 SSX24942.1 CCAG010017825 EZ422472 ADD18748.1 JXJN01009258 GDAI01000281 JAI17322.1 GAMC01011826 GAMC01011824 GAMC01011822 JAB94731.1 GEDC01001850 JAS35448.1 GECZ01023897 JAS45872.1 LBMM01003720 KMQ93205.1 GEBQ01013598 JAT26379.1 GL888818 EGI57898.1 KQ976662 KYM78551.1 DS235442 EEB15794.1 ADTU01014830 KZ288405 PBC26169.1 GL435242 EFN73689.1 JR052861 AEY61803.1 GL768221 EFZ11769.1 KQ414632 KOC67205.1 GL451202 EFN79748.1 KK107135 QOIP01000006 EZA58037.1 RLU21621.1 KQ979568 KYN20885.1 KQ982314 KYQ57907.1 KQ434936 KZC11907.1 GBYB01008176 JAG77943.1 KQ435922 KOX68540.1 KQ981864 KYN34251.1 GECU01020246 JAS87460.1 AJWK01017457 GECU01015779 JAS91927.1 GDHF01033223 GDHF01012808 GDHF01011986 GDHF01009473 GDHF01008081 JAI19091.1 JAI39506.1 JAI40328.1 JAI42841.1 JAI44233.1 GFDF01003163 JAV10921.1 ATLV01022881 KE525337 KFB48029.1 GFDF01003164 JAV10920.1 AAAB01008900 EAA09452.4 APCN01002982 AXCM01005203 CH916367 EDW01317.1 AAAB01006600 EAA02814.3 CH940648 KRF79938.1 CH902617 EDV42674.1 CH933808 EDW10410.1 GAPW01004045 JAC09553.1 CM000160 EDW95907.2 OUUW01000013 SPP88006.1 CM000070 EAL27682.4 AE014297 BT012450 AAF55556.1 AAF55557.1 AAN13772.1 AAN13773.1 AAS93721.1 BT010013 AAQ22482.1 AXCN02000947 BT001886 AAN71660.1 CH964272 EDW84536.1 EDW61360.2 PYGN01000284 PSN49340.1 GDHC01020290 JAP98338.1
GDQN01002510 JAT87243.1 JAT88544.1 NWSH01001566 PCG70840.1 PCG70841.1 GAIX01000124 JAA92436.1 KQ459185 KPJ03306.1 KQ461073 KPJ09615.1 ODYU01013106 SOQ59791.1 AGBW02014665 OWR41255.1 KQ971352 EFA06631.1 NEVH01006994 PNF36090.1 PNF36088.1 KK853284 KDR08981.1 GEZM01004710 JAV96325.1 GALX01005889 JAB62577.1 APGK01028620 BT126880 KB740694 KB632300 AEE61842.1 ENN79461.1 ERL91750.1 GANO01003154 JAB56717.1 JRES01001049 KNC25973.1 JXUM01077808 KQ563028 KXJ74617.1 DS231870 EDS41009.1 CH477227 EAT47089.1 GFDL01015268 JAV19777.1 UFQT01000523 SSX24942.1 CCAG010017825 EZ422472 ADD18748.1 JXJN01009258 GDAI01000281 JAI17322.1 GAMC01011826 GAMC01011824 GAMC01011822 JAB94731.1 GEDC01001850 JAS35448.1 GECZ01023897 JAS45872.1 LBMM01003720 KMQ93205.1 GEBQ01013598 JAT26379.1 GL888818 EGI57898.1 KQ976662 KYM78551.1 DS235442 EEB15794.1 ADTU01014830 KZ288405 PBC26169.1 GL435242 EFN73689.1 JR052861 AEY61803.1 GL768221 EFZ11769.1 KQ414632 KOC67205.1 GL451202 EFN79748.1 KK107135 QOIP01000006 EZA58037.1 RLU21621.1 KQ979568 KYN20885.1 KQ982314 KYQ57907.1 KQ434936 KZC11907.1 GBYB01008176 JAG77943.1 KQ435922 KOX68540.1 KQ981864 KYN34251.1 GECU01020246 JAS87460.1 AJWK01017457 GECU01015779 JAS91927.1 GDHF01033223 GDHF01012808 GDHF01011986 GDHF01009473 GDHF01008081 JAI19091.1 JAI39506.1 JAI40328.1 JAI42841.1 JAI44233.1 GFDF01003163 JAV10921.1 ATLV01022881 KE525337 KFB48029.1 GFDF01003164 JAV10920.1 AAAB01008900 EAA09452.4 APCN01002982 AXCM01005203 CH916367 EDW01317.1 AAAB01006600 EAA02814.3 CH940648 KRF79938.1 CH902617 EDV42674.1 CH933808 EDW10410.1 GAPW01004045 JAC09553.1 CM000160 EDW95907.2 OUUW01000013 SPP88006.1 CM000070 EAL27682.4 AE014297 BT012450 AAF55556.1 AAF55557.1 AAN13772.1 AAN13773.1 AAS93721.1 BT010013 AAQ22482.1 AXCN02000947 BT001886 AAN71660.1 CH964272 EDW84536.1 EDW61360.2 PYGN01000284 PSN49340.1 GDHC01020290 JAP98338.1
Proteomes
UP000005204
UP000283053
UP000037510
UP000218220
UP000053268
UP000053240
+ More
UP000007151 UP000007266 UP000235965 UP000027135 UP000192223 UP000019118 UP000030742 UP000037069 UP000069940 UP000249989 UP000002320 UP000008820 UP000092445 UP000092444 UP000092443 UP000092460 UP000091820 UP000095300 UP000078200 UP000095301 UP000036403 UP000007755 UP000078540 UP000009046 UP000005205 UP000242457 UP000000311 UP000005203 UP000053825 UP000008237 UP000053097 UP000279307 UP000078492 UP000075809 UP000076502 UP000053105 UP000078541 UP000092461 UP000075881 UP000075903 UP000030765 UP000075880 UP000075882 UP000007062 UP000075840 UP000075902 UP000002358 UP000076407 UP000075883 UP000001070 UP000075920 UP000075901 UP000076408 UP000008792 UP000007801 UP000009192 UP000192221 UP000002282 UP000075885 UP000075884 UP000268350 UP000001819 UP000000803 UP000075886 UP000007798 UP000245037
UP000007151 UP000007266 UP000235965 UP000027135 UP000192223 UP000019118 UP000030742 UP000037069 UP000069940 UP000249989 UP000002320 UP000008820 UP000092445 UP000092444 UP000092443 UP000092460 UP000091820 UP000095300 UP000078200 UP000095301 UP000036403 UP000007755 UP000078540 UP000009046 UP000005205 UP000242457 UP000000311 UP000005203 UP000053825 UP000008237 UP000053097 UP000279307 UP000078492 UP000075809 UP000076502 UP000053105 UP000078541 UP000092461 UP000075881 UP000075903 UP000030765 UP000075880 UP000075882 UP000007062 UP000075840 UP000075902 UP000002358 UP000076407 UP000075883 UP000001070 UP000075920 UP000075901 UP000076408 UP000008792 UP000007801 UP000009192 UP000192221 UP000002282 UP000075885 UP000075884 UP000268350 UP000001819 UP000000803 UP000075886 UP000007798 UP000245037
Interpro
ProteinModelPortal
H9J3D6
A0A3S2NSW2
A0A0L7LUU8
A0A1E1WJR3
A0A2A4JFT3
S4PIP3
+ More
A0A194QIM7 A0A194QXD2 A0A2H1X382 A0A212EIF2 D6WSE2 A0A2J7R5I9 A0A2J7R5I2 A0A067QJ65 A0A1W4X2Z6 A0A1Y1NI35 V5GLU9 J3JUG7 U5ESQ3 A0A0L0C0X5 A0A182GP49 B0WAB9 Q17K96 A0A1Q3EWT2 A0A336M3T8 A0A1B0A2D7 D3TM20 A0A1A9Y0Y7 A0A1B0B6X7 A0A1A9WD88 A0A1I8Q3R2 A0A1A9VFA3 A0A0K8TSI0 W8BMY0 A0A1B6EBY4 A0A1B6F6L5 A0A1I8MAC3 A0A0J7NL68 A0A1B6LS32 F4X6W6 A0A195B247 E0VQY8 A0A158NFX6 A0A2A3E4S4 E1ZYZ9 A0A088A6G8 V9IKZ2 E9J652 A0A0L7R8K7 E2BX29 A0A026WPQ2 A0A195E6R4 A0A151XC35 A0A154PJA8 A0A1A9X4T7 A0A0C9RMH0 A0A0M8ZPM1 A0A195F1J6 A0A1B6IKK8 A0A1B0CLM9 A0A1B6IYB4 A0A182KA64 A0A182V0C5 A0A0K8TXS4 A0A1L8DWT6 A0A084WCT5 A0A1L8DWS9 A0A182IVR9 A0A182LB61 Q7Q9T2 A0A182HPD5 A0A182TRE1 K7ILS7 A0A182X0W7 A0A182M4V5 B4J8Q0 Q7QLK2 A0A182WMB6 A0A182STZ8 A0A1A9V274 A0A182YJ16 A0A0Q9W541 B3LW96 B4KRI4 A0A1W4U8C8 A0A023EK93 B4PLX5 A0A182NZQ7 A0A1B0AF92 A0A182NLX0 A0A3B0KQ92 Q299M1 Q9VE72 Q7YU34 A0A182Q8V8 Q8IG98 B4NH39 B4LQ75 A0A2P8YYL4 A0A146KQP8
A0A194QIM7 A0A194QXD2 A0A2H1X382 A0A212EIF2 D6WSE2 A0A2J7R5I9 A0A2J7R5I2 A0A067QJ65 A0A1W4X2Z6 A0A1Y1NI35 V5GLU9 J3JUG7 U5ESQ3 A0A0L0C0X5 A0A182GP49 B0WAB9 Q17K96 A0A1Q3EWT2 A0A336M3T8 A0A1B0A2D7 D3TM20 A0A1A9Y0Y7 A0A1B0B6X7 A0A1A9WD88 A0A1I8Q3R2 A0A1A9VFA3 A0A0K8TSI0 W8BMY0 A0A1B6EBY4 A0A1B6F6L5 A0A1I8MAC3 A0A0J7NL68 A0A1B6LS32 F4X6W6 A0A195B247 E0VQY8 A0A158NFX6 A0A2A3E4S4 E1ZYZ9 A0A088A6G8 V9IKZ2 E9J652 A0A0L7R8K7 E2BX29 A0A026WPQ2 A0A195E6R4 A0A151XC35 A0A154PJA8 A0A1A9X4T7 A0A0C9RMH0 A0A0M8ZPM1 A0A195F1J6 A0A1B6IKK8 A0A1B0CLM9 A0A1B6IYB4 A0A182KA64 A0A182V0C5 A0A0K8TXS4 A0A1L8DWT6 A0A084WCT5 A0A1L8DWS9 A0A182IVR9 A0A182LB61 Q7Q9T2 A0A182HPD5 A0A182TRE1 K7ILS7 A0A182X0W7 A0A182M4V5 B4J8Q0 Q7QLK2 A0A182WMB6 A0A182STZ8 A0A1A9V274 A0A182YJ16 A0A0Q9W541 B3LW96 B4KRI4 A0A1W4U8C8 A0A023EK93 B4PLX5 A0A182NZQ7 A0A1B0AF92 A0A182NLX0 A0A3B0KQ92 Q299M1 Q9VE72 Q7YU34 A0A182Q8V8 Q8IG98 B4NH39 B4LQ75 A0A2P8YYL4 A0A146KQP8
PDB
6CKX
E-value=5.78122e-13,
Score=177
Ontologies
PANTHER
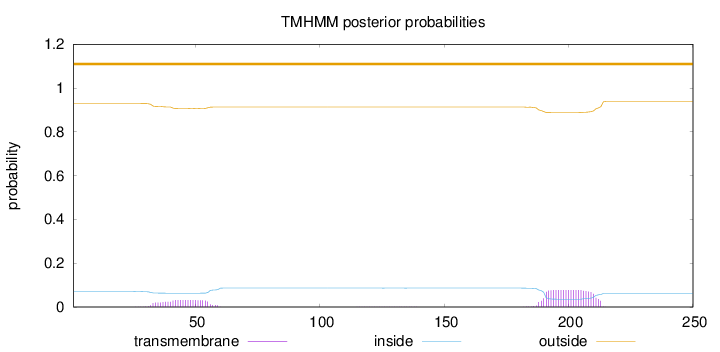
Topology
Length:
250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.39412
Exp number, first 60 AAs:
0.68084
Total prob of N-in:
0.07060
outside
1 - 250
Population Genetic Test Statistics
Pi
148.700458
Theta
156.129203
Tajima's D
-0.235404
CLR
124.363948
CSRT
0.308884555772211
Interpretation
Uncertain
