Gene
KWMTBOMO11576
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.708
Sequence
CDS
ATGGTAAACAGGTCAGCATTGTCTTACCGGGAGTCTGCCCGTCTGGCGTGCGCAGGCGTGGCACACGTCGTCACAAGGATCCGACGGCTGGGACTCGAGGTGGCGCTCGACAAAACCCAGGCCCTGTTGTTTCACGGGCCGGGACGAGCGCCGCCTCTGGGTGCCCACCTCGTGATCGGAGGCGTCCGCGTCGGGGTCGGGGTGACCGGTCTTCGGTACCTCGGCCTTGAACTGGATGGTCGGTGGAACTTCCGCGCTCACTTTGCGAAGTTGGGCCCTCGACTGATGGCGACGGCCGGCTCTTTGAGCCGGCTGCTTCCAAACGTCGGGGGTCCCGATCAGGTGGCGCGCCGCCTCTACATGGGGGTGGTGCGGTCGATGGCACTATACGGCGCGCCCGTATGGTGCCACGCCCTGACCCGCCAGAACGTCGCCGCGCTGCGACGTCCGCAGCGCGCGATCACGGTCAGGGCGATCCGAGGATACCGCACCGTCTCCTTCGAGGCGGCGTGCTTGCTCGCCGGGGCGCCACCCTGGGACCTGGAGGCGGAGGCGCTCGCTGCCGATTACCGGCTCCAATCTCGGCGGTCCGTGCTGGAGGCGTGGTCTCGCCGCTTGGCGGACCCGTCGGCCGGCCTCCGTACCGTCGAGGCGGTCCGTCCGGTCCTCGCGGACTGGGTGGGCCGCGACCGCGGATGCCTCACCTTCCGGCTGACGCAGATGTTGACCGGCCATGGGTGTTTTGGCCGGTACCTCTTTAAGATAGCCGGAAGGGAACCGACGGCGCAGTGCCACCACTGCGCGGACCGCGACGAGGAAGACACAGCGGAACACACGCTGGCGCGTTGCTCTGGATTCGACGAGCAACGCGCCGCCCTCGTCGCGGTCATTGGAGAGGACCTCTCGCTGCCGCGCGTCGTGGCTACGATGCTCGGCAGCGACGCGTCCTGGAAGGCGATGCTCGACTTCTGCGAGTCCACCATCTCGCAGAAGGAGGCGGCGGAGCGAGAGAGGGAGAGCTCTTCCCTTTCCGCACCGATCCGTCGCCGTCGAGCCGGGGGCCGGAGCACAGTGGGTCGATACTGTGATTTTAGTGACTTAGGGACCAACTTTGTTTTTTTACATTTTTCTTTATGTAGATAG
Protein
MVNRSALSYRESARLACAGVAHVVTRIRRLGLEVALDKTQALLFHGPGRAPPLGAHLVIGGVRVGVGVTGLRYLGLELDGRWNFRAHFAKLGPRLMATAGSLSRLLPNVGGPDQVARRLYMGVVRSMALYGAPVWCHALTRQNVAALRRPQRAITVRAIRGYRTVSFEAACLLAGAPPWDLEAEALAADYRLQSRRSVLEAWSRRLADPSAGLRTVEAVRPVLADWVGRDRGCLTFRLTQMLTGHGCFGRYLFKIAGREPTAQCHHCADRDEEDTAEHTLARCSGFDEQRAALVAVIGEDLSLPRVVATMLGSDASWKAMLDFCESTISQKEAAERERESSSLSAPIRRRRAGGRSTVGRYCDFSDLGTNFVFLHFSLCR
Summary
Uniprot
Q5KTM7
Q868Q4
A0A2W1B333
Q8MY27
Q8MY31
Q8MY33
+ More
Q8MY35 A0A2W1BDU5 A0A3S2N998 A0A2W1C3J5 A0A0J7N1D9 O01419 A0A0J7N7H9 A0A0J7KM11 A0A0J7MY07 A0A0J7KCQ3 E2BVR6 A0A0J7KIY5 A0A0J7JZ13 A0A0J7JZF7 A0A0J7KIW1 A0A0J7KJH4 A0A0J7KQK3 A0A0J7KJG6 A0A194R5D3 A0A0J7N7S8 A0A3L8DDV9 A0A0J7KS67 A0A0J7KL06 A0A0J7KIM6 A0A0J7KC11 A0A0J7KM81 A0A0J7KFG9 A0A0J7KLT9 A0A0J7K3R6 A0A3L8D2G3 A0A0J7KF37 A0A0J7KMU1 A0A0J7KPZ3 A0A0J7KPR0 A0A0J7K3F1 A0A0J7KYD2 A0A0J7NMN5 A0A0J7KRB8 A0A3S2LNH8 A0A0J7KP92 A0A0J7KWU5 A0A3L8DIL1 A0A0J7KLH8 A0A0J7NAV8 A0A0J7JXX4 A0A0J7K3R2 A0A0J7KGJ6 A0A0J7KF40 A0A1B6HLL2 A0A0J7NDI6 A0A0J7JYP2 A0A0J7KCZ9 A0A2S2QW03 A0A0J7KW81 A0A2S2PEZ7 J9KHF7 J9KV04 V5I8F3 A0A0J7N0R3 A0A0J7KFH2 A0A2S2N8I2
Q8MY35 A0A2W1BDU5 A0A3S2N998 A0A2W1C3J5 A0A0J7N1D9 O01419 A0A0J7N7H9 A0A0J7KM11 A0A0J7MY07 A0A0J7KCQ3 E2BVR6 A0A0J7KIY5 A0A0J7JZ13 A0A0J7JZF7 A0A0J7KIW1 A0A0J7KJH4 A0A0J7KQK3 A0A0J7KJG6 A0A194R5D3 A0A0J7N7S8 A0A3L8DDV9 A0A0J7KS67 A0A0J7KL06 A0A0J7KIM6 A0A0J7KC11 A0A0J7KM81 A0A0J7KFG9 A0A0J7KLT9 A0A0J7K3R6 A0A3L8D2G3 A0A0J7KF37 A0A0J7KMU1 A0A0J7KPZ3 A0A0J7KPR0 A0A0J7K3F1 A0A0J7KYD2 A0A0J7NMN5 A0A0J7KRB8 A0A3S2LNH8 A0A0J7KP92 A0A0J7KWU5 A0A3L8DIL1 A0A0J7KLH8 A0A0J7NAV8 A0A0J7JXX4 A0A0J7K3R2 A0A0J7KGJ6 A0A0J7KF40 A0A1B6HLL2 A0A0J7NDI6 A0A0J7JYP2 A0A0J7KCZ9 A0A2S2QW03 A0A0J7KW81 A0A2S2PEZ7 J9KHF7 J9KV04 V5I8F3 A0A0J7N0R3 A0A0J7KFH2 A0A2S2N8I2
EMBL
AB126050
BAD86650.1
AB090825
BAC57926.1
KZ150386
PZC71062.1
+ More
AB078935 BAC06462.1 AB078931 BAC06456.1 AB078930 BAC06454.1 AB078929 BAC06452.1 KZ150308 PZC71477.1 RSAL01000481 RVE41577.1 KZ149896 PZC78733.1 LBMM01012027 KMQ86470.1 D85594 BAA19776.1 LBMM01008861 KMQ88580.1 LBMM01005707 KMQ91279.1 LBMM01014132 KMQ85305.1 LBMM01009494 KMQ88097.1 GL450953 EFN80213.1 LBMM01006954 KMQ90166.1 LBMM01019839 KMQ83438.1 LBMM01019838 KMQ83439.1 LBMM01006725 KMQ90373.1 LBMM01006657 KMQ90417.1 LBMM01004272 KMQ92618.1 LBMM01006671 KMQ90407.1 KQ460685 KPJ12887.1 LBMM01008682 KMQ88705.1 QOIP01000010 RLU18098.1 LBMM01003679 KMQ93242.1 LBMM01006063 KMQ90967.1 LBMM01007036 KMQ90099.1 LBMM01009738 KMQ87908.1 LBMM01005634 KMQ91349.1 LBMM01008349 KMQ88951.1 LBMM01005733 KMQ91252.1 LBMM01015018 KMQ84954.1 QOIP01000031 RLU14697.1 LBMM01008448 KMQ88867.1 LBMM01005216 KMQ91703.1 LBMM01004505 KMQ92346.1 LBMM01004634 KMQ92224.1 LBMM01015025 KMQ84953.1 LBMM01002052 KMQ95336.1 LBMM01003245 KMQ93755.1 LBMM01003988 KMQ92922.1 RSAL01003504 RVE40273.1 LBMM01004697 KMQ92167.1 LBMM01002480 KMQ94776.1 QOIP01000007 RLU20280.1 LBMM01005948 KMQ91066.1 LBMM01007425 KMQ89740.1 LBMM01021647 KMQ83033.1 LBMM01014956 KMQ84982.1 LBMM01007659 KMQ89538.1 LBMM01008422 KMQ88894.1 GECU01032129 JAS75577.1 LBMM01006496 KMQ90570.1 LBMM01020849 KMQ83202.1 LBMM01009527 LBMM01009515 KMQ88059.1 KMQ88067.1 GGMS01012744 MBY81947.1 KMQ94772.1 GGMR01015159 MBY27778.1 ABLF02014530 ABLF02018586 ABLF02058000 ABLF02004674 ABLF02004675 ABLF02004676 ABLF02011637 GALX01004785 JAB63681.1 LBMM01012412 KMQ86235.1 LBMM01008278 KMQ89009.1 GGMR01000779 MBY13398.1
AB078935 BAC06462.1 AB078931 BAC06456.1 AB078930 BAC06454.1 AB078929 BAC06452.1 KZ150308 PZC71477.1 RSAL01000481 RVE41577.1 KZ149896 PZC78733.1 LBMM01012027 KMQ86470.1 D85594 BAA19776.1 LBMM01008861 KMQ88580.1 LBMM01005707 KMQ91279.1 LBMM01014132 KMQ85305.1 LBMM01009494 KMQ88097.1 GL450953 EFN80213.1 LBMM01006954 KMQ90166.1 LBMM01019839 KMQ83438.1 LBMM01019838 KMQ83439.1 LBMM01006725 KMQ90373.1 LBMM01006657 KMQ90417.1 LBMM01004272 KMQ92618.1 LBMM01006671 KMQ90407.1 KQ460685 KPJ12887.1 LBMM01008682 KMQ88705.1 QOIP01000010 RLU18098.1 LBMM01003679 KMQ93242.1 LBMM01006063 KMQ90967.1 LBMM01007036 KMQ90099.1 LBMM01009738 KMQ87908.1 LBMM01005634 KMQ91349.1 LBMM01008349 KMQ88951.1 LBMM01005733 KMQ91252.1 LBMM01015018 KMQ84954.1 QOIP01000031 RLU14697.1 LBMM01008448 KMQ88867.1 LBMM01005216 KMQ91703.1 LBMM01004505 KMQ92346.1 LBMM01004634 KMQ92224.1 LBMM01015025 KMQ84953.1 LBMM01002052 KMQ95336.1 LBMM01003245 KMQ93755.1 LBMM01003988 KMQ92922.1 RSAL01003504 RVE40273.1 LBMM01004697 KMQ92167.1 LBMM01002480 KMQ94776.1 QOIP01000007 RLU20280.1 LBMM01005948 KMQ91066.1 LBMM01007425 KMQ89740.1 LBMM01021647 KMQ83033.1 LBMM01014956 KMQ84982.1 LBMM01007659 KMQ89538.1 LBMM01008422 KMQ88894.1 GECU01032129 JAS75577.1 LBMM01006496 KMQ90570.1 LBMM01020849 KMQ83202.1 LBMM01009527 LBMM01009515 KMQ88059.1 KMQ88067.1 GGMS01012744 MBY81947.1 KMQ94772.1 GGMR01015159 MBY27778.1 ABLF02014530 ABLF02018586 ABLF02058000 ABLF02004674 ABLF02004675 ABLF02004676 ABLF02011637 GALX01004785 JAB63681.1 LBMM01012412 KMQ86235.1 LBMM01008278 KMQ89009.1 GGMR01000779 MBY13398.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
Q5KTM7
Q868Q4
A0A2W1B333
Q8MY27
Q8MY31
Q8MY33
+ More
Q8MY35 A0A2W1BDU5 A0A3S2N998 A0A2W1C3J5 A0A0J7N1D9 O01419 A0A0J7N7H9 A0A0J7KM11 A0A0J7MY07 A0A0J7KCQ3 E2BVR6 A0A0J7KIY5 A0A0J7JZ13 A0A0J7JZF7 A0A0J7KIW1 A0A0J7KJH4 A0A0J7KQK3 A0A0J7KJG6 A0A194R5D3 A0A0J7N7S8 A0A3L8DDV9 A0A0J7KS67 A0A0J7KL06 A0A0J7KIM6 A0A0J7KC11 A0A0J7KM81 A0A0J7KFG9 A0A0J7KLT9 A0A0J7K3R6 A0A3L8D2G3 A0A0J7KF37 A0A0J7KMU1 A0A0J7KPZ3 A0A0J7KPR0 A0A0J7K3F1 A0A0J7KYD2 A0A0J7NMN5 A0A0J7KRB8 A0A3S2LNH8 A0A0J7KP92 A0A0J7KWU5 A0A3L8DIL1 A0A0J7KLH8 A0A0J7NAV8 A0A0J7JXX4 A0A0J7K3R2 A0A0J7KGJ6 A0A0J7KF40 A0A1B6HLL2 A0A0J7NDI6 A0A0J7JYP2 A0A0J7KCZ9 A0A2S2QW03 A0A0J7KW81 A0A2S2PEZ7 J9KHF7 J9KV04 V5I8F3 A0A0J7N0R3 A0A0J7KFH2 A0A2S2N8I2
Q8MY35 A0A2W1BDU5 A0A3S2N998 A0A2W1C3J5 A0A0J7N1D9 O01419 A0A0J7N7H9 A0A0J7KM11 A0A0J7MY07 A0A0J7KCQ3 E2BVR6 A0A0J7KIY5 A0A0J7JZ13 A0A0J7JZF7 A0A0J7KIW1 A0A0J7KJH4 A0A0J7KQK3 A0A0J7KJG6 A0A194R5D3 A0A0J7N7S8 A0A3L8DDV9 A0A0J7KS67 A0A0J7KL06 A0A0J7KIM6 A0A0J7KC11 A0A0J7KM81 A0A0J7KFG9 A0A0J7KLT9 A0A0J7K3R6 A0A3L8D2G3 A0A0J7KF37 A0A0J7KMU1 A0A0J7KPZ3 A0A0J7KPR0 A0A0J7K3F1 A0A0J7KYD2 A0A0J7NMN5 A0A0J7KRB8 A0A3S2LNH8 A0A0J7KP92 A0A0J7KWU5 A0A3L8DIL1 A0A0J7KLH8 A0A0J7NAV8 A0A0J7JXX4 A0A0J7K3R2 A0A0J7KGJ6 A0A0J7KF40 A0A1B6HLL2 A0A0J7NDI6 A0A0J7JYP2 A0A0J7KCZ9 A0A2S2QW03 A0A0J7KW81 A0A2S2PEZ7 J9KHF7 J9KV04 V5I8F3 A0A0J7N0R3 A0A0J7KFH2 A0A2S2N8I2
Ontologies
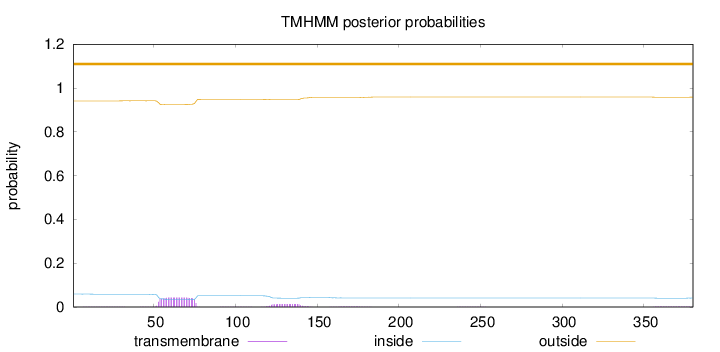
Topology
Length:
380
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.33022
Exp number, first 60 AAs:
0.33316
Total prob of N-in:
0.05926
outside
1 - 380
Population Genetic Test Statistics
Pi
20.370481
Theta
21.419416
Tajima's D
1.235425
CLR
0.225622
CSRT
0.725663716814159
Interpretation
Uncertain
