Gene
KWMTBOMO11572 Validated by peptides from experiments
Annotation
salivary_secreted_ribonuclease_[Bombyx_mori]
Full name
Ribonuclease kappa
+ More
Ribonuclease kappa-B
Ribonuclease kappa-B
Alternative Name
Cc RNase
Location in the cell
PlasmaMembrane Reliability : 2.567
Sequence
CDS
ATGGGCTGCAAACTGTGTGGACCAAAACTGTCGCTTTGTGGCCTAGTTTTGAGCGTATGGGGCATCATTCAGTTGACGTTGATGGGCGTCTTTTATTATATTCGAGCTGTCGCTCTGCTGGAGGATCTACCATTTGATGAGAAAAATCCGCCTCATTCTATCGAAGACTTCGTCATTGAAGTGGAAAAAGGATATACCCTGAATGCACAAAATTGTTGGATTGCGGCATTGTTGTACTTGATTACACTGGTTGTATCTGGACATCAATTTTGGCTGAATAATCGTTCCTCAGTTAGTATGTAA
Protein
MGCKLCGPKLSLCGLVLSVWGIIQLTLMGVFYYIRAVALLEDLPFDEKNPPHSIEDFVIEVEKGYTLNAQNCWIAALLYLITLVVSGHQFWLNNRSSVSM
Summary
Description
Endoribonuclease.
Endoribonuclease which displays activity against poly(C) and poly(U) synthetic substrates, as well as rRNA.
Endoribonuclease which displays activity against poly(C) and poly(U) synthetic substrates, as well as rRNA.
Similarity
Belongs to the RNase K family.
Keywords
Complete proteome
Endonuclease
Hydrolase
Membrane
Nuclease
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Ribonuclease kappa
Uniprot
Q1HQB1
A0A2A4K6A1
B6A8I2
A0A212EX66
A0A2H1WE14
A0A3S2LZJ0
+ More
S4NGP4 I4DK55 H9J3K4 A0A194QWE4 Q1W2C7 A0A1S3DHU8 N6UR57 A0A1W4WMP2 J3JV44 D6WRM1 A0A2P8ZM12 E2AJ54 A0A026WB54 A0A151JVL1 A0A158P3Y3 E2BGG9 A0A067RJH1 F4WKU1 A0A232ERB4 K7IS82 A0A0M8ZW62 U5ESR9 A0A151J8U3 A0A154PGH3 B4GBJ0 A0A1Q3FKA5 B0VZA4 B5E0W0 A0A0B4KEQ9 B4ISK9 B4IMC2 B3NKK0 B4NSS8 Q8MSF5 A0A1W4VF31 A0A088AJC6 A0A2A3E8M6 T1D5L0 Q7Z0Q2 B4MJ04 A0A084VZ21 A0A023EEH7 A0A1J1I5D4 A0A0L0C314 B3MBR6 A0A182JZ97 A0A182WC98 A0A2P8YJQ9 B4J9F1 Q1HRK4 A0A182WTU5 Q7Q5K0 A0A182HI42 R4WRC1 A0A182R5J7 A0A1B0GI92 A0A1L8D8Q5 R4G495 A0A182Q8H1 A0A2M3Z1G3 A0A182PET5 A0A2M4ADV5 A0A2M4C4H2 W5JR74 A0A182FEL0 D3PFQ2 A0A2M4A704 A6YPE7 J9KM02 A0A0V0G565 A0A023FB88 A0A336M0C3 A0A0M4ECV7 A0A069DVD2 A0A224Y1Z2 B4LPH6 E9G2F9 A0A195BKP1 A0A1B0B5C1 A0A1B0A3N9 D3TM88 A0A0N8B6T8 A0A0N8A335 A0A0P5TMI1 A0A0P5J892 A0A0N8AHA3 A0A0P4YYM6 A0A0L7RBZ3 D3PHH8 C4N185 A0A1I8N878 A0A1L8EEZ5 A0A1I8PAY8 A0A1D2NAB6
S4NGP4 I4DK55 H9J3K4 A0A194QWE4 Q1W2C7 A0A1S3DHU8 N6UR57 A0A1W4WMP2 J3JV44 D6WRM1 A0A2P8ZM12 E2AJ54 A0A026WB54 A0A151JVL1 A0A158P3Y3 E2BGG9 A0A067RJH1 F4WKU1 A0A232ERB4 K7IS82 A0A0M8ZW62 U5ESR9 A0A151J8U3 A0A154PGH3 B4GBJ0 A0A1Q3FKA5 B0VZA4 B5E0W0 A0A0B4KEQ9 B4ISK9 B4IMC2 B3NKK0 B4NSS8 Q8MSF5 A0A1W4VF31 A0A088AJC6 A0A2A3E8M6 T1D5L0 Q7Z0Q2 B4MJ04 A0A084VZ21 A0A023EEH7 A0A1J1I5D4 A0A0L0C314 B3MBR6 A0A182JZ97 A0A182WC98 A0A2P8YJQ9 B4J9F1 Q1HRK4 A0A182WTU5 Q7Q5K0 A0A182HI42 R4WRC1 A0A182R5J7 A0A1B0GI92 A0A1L8D8Q5 R4G495 A0A182Q8H1 A0A2M3Z1G3 A0A182PET5 A0A2M4ADV5 A0A2M4C4H2 W5JR74 A0A182FEL0 D3PFQ2 A0A2M4A704 A6YPE7 J9KM02 A0A0V0G565 A0A023FB88 A0A336M0C3 A0A0M4ECV7 A0A069DVD2 A0A224Y1Z2 B4LPH6 E9G2F9 A0A195BKP1 A0A1B0B5C1 A0A1B0A3N9 D3TM88 A0A0N8B6T8 A0A0N8A335 A0A0P5TMI1 A0A0P5J892 A0A0N8AHA3 A0A0P4YYM6 A0A0L7RBZ3 D3PHH8 C4N185 A0A1I8N878 A0A1L8EEZ5 A0A1I8PAY8 A0A1D2NAB6
EC Number
3.1.-.-
Pubmed
22118469
23622113
22651552
19121390
26354079
23537049
+ More
22516182 18362917 19820115 29403074 20798317 24508170 30249741 21347285 24845553 21719571 28648823 20075255 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 12537569 24330624 12799437 24438588 24945155 26483478 26108605 17204158 17510324 12364791 14747013 17210077 23691247 20920257 23761445 18207082 25474469 26334808 21292972 20353571 19576987 25315136 27289101
22516182 18362917 19820115 29403074 20798317 24508170 30249741 21347285 24845553 21719571 28648823 20075255 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 12537569 24330624 12799437 24438588 24945155 26483478 26108605 17204158 17510324 12364791 14747013 17210077 23691247 20920257 23761445 18207082 25474469 26334808 21292972 20353571 19576987 25315136 27289101
EMBL
DQ443141
ABF51230.1
NWSH01000111
PCG79448.1
DQ875242
KP141855
+ More
ABK29484.1 AJW81856.1 AGBW02011827 OWR46073.1 ODYU01008049 SOQ51325.1 RSAL01000096 RVE47720.1 GAIX01014664 JAA77896.1 AK401673 BAM18295.1 BABH01021511 BABH01021512 KQ461073 KPJ09624.1 DQ445500 ABD98738.1 APGK01019947 KB740150 KB632173 ENN81242.1 ERL89567.1 BT127110 AEE62072.1 KQ971354 EFA05973.1 PYGN01000019 PSN57536.1 GL439967 EFN66494.1 KK107309 QOIP01000011 EZA52901.1 RLU16393.1 KQ981701 KYN37494.1 ADTU01008610 GL448181 EFN85218.1 KK852473 KDR23123.1 GL888206 EGI65183.1 NNAY01002629 OXU20900.1 KQ435851 KOX70789.1 GANO01002235 JAB57636.1 KQ979480 KYN21280.1 KQ434899 KZC10943.1 CH479181 EDW31285.1 GFDL01007103 JAV27942.1 DS231813 EDS25706.1 CM000071 EDY68670.2 AE013599 AGB93234.1 CH891577 EDW99364.1 CH480967 EDW45600.1 CH954179 EDV54304.1 CH982438 CM002911 EDX15260.1 KMY91506.1 AY118852 KZ288338 PBC27814.1 GALA01000458 JAA94394.1 AJ441124 AJ874688 AJ874689 AJ874690 CH963719 EDW72093.1 ATLV01018587 KE525239 KFB43215.1 JXUM01026157 JXUM01026158 GAPW01006554 GAPW01006553 GAPW01006551 KQ560746 JAC07044.1 KXJ81029.1 CVRI01000042 CRK95501.1 JRES01000960 KNC26715.1 CH902619 EDV38212.1 PYGN01000545 PSN44490.1 CH916367 EDW01432.1 DQ440090 CH477273 ABF18123.1 EAT45204.1 AAAB01008960 EAA11171.3 APCN01005875 AK417222 BAN20437.1 AJWK01013575 GFDF01011251 JAV02833.1 ACPB03001659 GAHY01000956 JAA76554.1 AXCN02000886 GGFM01001602 MBW22353.1 GGFK01005642 MBW38963.1 GGFJ01011095 MBW60236.1 ADMH02000413 ETN66636.1 BT120458 BT121328 HACA01003756 ADD24098.1 ADD38258.1 CDW21117.1 GGFK01003087 MBW36408.1 EF638958 ABR27843.1 ABLF02016577 ABLF02016578 GECL01003300 JAP02824.1 GBBI01000087 JAC18625.1 UFQS01000381 UFQT01000381 SSX03411.1 SSX23776.1 CP012524 ALC40676.1 GBGD01003620 JAC85269.1 GFTR01001384 JAW15042.1 CH940648 EDW61235.1 GL732530 EFX86315.1 KQ976453 KYM85339.1 JXJN01008672 CCAG010007832 EZ422540 EZ422541 ADD18816.1 ADD18817.1 GDIQ01207155 JAK44570.1 GDIP01187257 JAJ36145.1 GDIP01124264 JAL79450.1 GDIP01152729 GDIQ01209706 JAJ70673.1 JAK42019.1 GDIP01147513 JAJ75889.1 GDIP01221003 JAJ02399.1 KQ414617 KOC68330.1 BT121084 ADD38014.1 EZ048858 ACN69150.1 GFDG01001517 JAV17282.1 LJIJ01000120 ODN02203.1
ABK29484.1 AJW81856.1 AGBW02011827 OWR46073.1 ODYU01008049 SOQ51325.1 RSAL01000096 RVE47720.1 GAIX01014664 JAA77896.1 AK401673 BAM18295.1 BABH01021511 BABH01021512 KQ461073 KPJ09624.1 DQ445500 ABD98738.1 APGK01019947 KB740150 KB632173 ENN81242.1 ERL89567.1 BT127110 AEE62072.1 KQ971354 EFA05973.1 PYGN01000019 PSN57536.1 GL439967 EFN66494.1 KK107309 QOIP01000011 EZA52901.1 RLU16393.1 KQ981701 KYN37494.1 ADTU01008610 GL448181 EFN85218.1 KK852473 KDR23123.1 GL888206 EGI65183.1 NNAY01002629 OXU20900.1 KQ435851 KOX70789.1 GANO01002235 JAB57636.1 KQ979480 KYN21280.1 KQ434899 KZC10943.1 CH479181 EDW31285.1 GFDL01007103 JAV27942.1 DS231813 EDS25706.1 CM000071 EDY68670.2 AE013599 AGB93234.1 CH891577 EDW99364.1 CH480967 EDW45600.1 CH954179 EDV54304.1 CH982438 CM002911 EDX15260.1 KMY91506.1 AY118852 KZ288338 PBC27814.1 GALA01000458 JAA94394.1 AJ441124 AJ874688 AJ874689 AJ874690 CH963719 EDW72093.1 ATLV01018587 KE525239 KFB43215.1 JXUM01026157 JXUM01026158 GAPW01006554 GAPW01006553 GAPW01006551 KQ560746 JAC07044.1 KXJ81029.1 CVRI01000042 CRK95501.1 JRES01000960 KNC26715.1 CH902619 EDV38212.1 PYGN01000545 PSN44490.1 CH916367 EDW01432.1 DQ440090 CH477273 ABF18123.1 EAT45204.1 AAAB01008960 EAA11171.3 APCN01005875 AK417222 BAN20437.1 AJWK01013575 GFDF01011251 JAV02833.1 ACPB03001659 GAHY01000956 JAA76554.1 AXCN02000886 GGFM01001602 MBW22353.1 GGFK01005642 MBW38963.1 GGFJ01011095 MBW60236.1 ADMH02000413 ETN66636.1 BT120458 BT121328 HACA01003756 ADD24098.1 ADD38258.1 CDW21117.1 GGFK01003087 MBW36408.1 EF638958 ABR27843.1 ABLF02016577 ABLF02016578 GECL01003300 JAP02824.1 GBBI01000087 JAC18625.1 UFQS01000381 UFQT01000381 SSX03411.1 SSX23776.1 CP012524 ALC40676.1 GBGD01003620 JAC85269.1 GFTR01001384 JAW15042.1 CH940648 EDW61235.1 GL732530 EFX86315.1 KQ976453 KYM85339.1 JXJN01008672 CCAG010007832 EZ422540 EZ422541 ADD18816.1 ADD18817.1 GDIQ01207155 JAK44570.1 GDIP01187257 JAJ36145.1 GDIP01124264 JAL79450.1 GDIP01152729 GDIQ01209706 JAJ70673.1 JAK42019.1 GDIP01147513 JAJ75889.1 GDIP01221003 JAJ02399.1 KQ414617 KOC68330.1 BT121084 ADD38014.1 EZ048858 ACN69150.1 GFDG01001517 JAV17282.1 LJIJ01000120 ODN02203.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000005204
UP000053240
UP000079169
+ More
UP000019118 UP000030742 UP000192223 UP000007266 UP000245037 UP000000311 UP000053097 UP000279307 UP000078541 UP000005205 UP000008237 UP000027135 UP000007755 UP000215335 UP000002358 UP000053105 UP000078492 UP000076502 UP000008744 UP000002320 UP000001819 UP000000803 UP000002282 UP000001292 UP000008711 UP000000304 UP000192221 UP000005203 UP000242457 UP000007798 UP000030765 UP000069940 UP000249989 UP000183832 UP000037069 UP000007801 UP000075881 UP000075920 UP000001070 UP000008820 UP000076407 UP000007062 UP000075840 UP000075900 UP000092461 UP000015103 UP000075886 UP000075885 UP000000673 UP000069272 UP000007819 UP000092553 UP000008792 UP000000305 UP000078540 UP000092460 UP000092445 UP000092444 UP000053825 UP000095301 UP000095300 UP000094527
UP000019118 UP000030742 UP000192223 UP000007266 UP000245037 UP000000311 UP000053097 UP000279307 UP000078541 UP000005205 UP000008237 UP000027135 UP000007755 UP000215335 UP000002358 UP000053105 UP000078492 UP000076502 UP000008744 UP000002320 UP000001819 UP000000803 UP000002282 UP000001292 UP000008711 UP000000304 UP000192221 UP000005203 UP000242457 UP000007798 UP000030765 UP000069940 UP000249989 UP000183832 UP000037069 UP000007801 UP000075881 UP000075920 UP000001070 UP000008820 UP000076407 UP000007062 UP000075840 UP000075900 UP000092461 UP000015103 UP000075886 UP000075885 UP000000673 UP000069272 UP000007819 UP000092553 UP000008792 UP000000305 UP000078540 UP000092460 UP000092445 UP000092444 UP000053825 UP000095301 UP000095300 UP000094527
Interpro
IPR026770
RNase_K
ProteinModelPortal
Q1HQB1
A0A2A4K6A1
B6A8I2
A0A212EX66
A0A2H1WE14
A0A3S2LZJ0
+ More
S4NGP4 I4DK55 H9J3K4 A0A194QWE4 Q1W2C7 A0A1S3DHU8 N6UR57 A0A1W4WMP2 J3JV44 D6WRM1 A0A2P8ZM12 E2AJ54 A0A026WB54 A0A151JVL1 A0A158P3Y3 E2BGG9 A0A067RJH1 F4WKU1 A0A232ERB4 K7IS82 A0A0M8ZW62 U5ESR9 A0A151J8U3 A0A154PGH3 B4GBJ0 A0A1Q3FKA5 B0VZA4 B5E0W0 A0A0B4KEQ9 B4ISK9 B4IMC2 B3NKK0 B4NSS8 Q8MSF5 A0A1W4VF31 A0A088AJC6 A0A2A3E8M6 T1D5L0 Q7Z0Q2 B4MJ04 A0A084VZ21 A0A023EEH7 A0A1J1I5D4 A0A0L0C314 B3MBR6 A0A182JZ97 A0A182WC98 A0A2P8YJQ9 B4J9F1 Q1HRK4 A0A182WTU5 Q7Q5K0 A0A182HI42 R4WRC1 A0A182R5J7 A0A1B0GI92 A0A1L8D8Q5 R4G495 A0A182Q8H1 A0A2M3Z1G3 A0A182PET5 A0A2M4ADV5 A0A2M4C4H2 W5JR74 A0A182FEL0 D3PFQ2 A0A2M4A704 A6YPE7 J9KM02 A0A0V0G565 A0A023FB88 A0A336M0C3 A0A0M4ECV7 A0A069DVD2 A0A224Y1Z2 B4LPH6 E9G2F9 A0A195BKP1 A0A1B0B5C1 A0A1B0A3N9 D3TM88 A0A0N8B6T8 A0A0N8A335 A0A0P5TMI1 A0A0P5J892 A0A0N8AHA3 A0A0P4YYM6 A0A0L7RBZ3 D3PHH8 C4N185 A0A1I8N878 A0A1L8EEZ5 A0A1I8PAY8 A0A1D2NAB6
S4NGP4 I4DK55 H9J3K4 A0A194QWE4 Q1W2C7 A0A1S3DHU8 N6UR57 A0A1W4WMP2 J3JV44 D6WRM1 A0A2P8ZM12 E2AJ54 A0A026WB54 A0A151JVL1 A0A158P3Y3 E2BGG9 A0A067RJH1 F4WKU1 A0A232ERB4 K7IS82 A0A0M8ZW62 U5ESR9 A0A151J8U3 A0A154PGH3 B4GBJ0 A0A1Q3FKA5 B0VZA4 B5E0W0 A0A0B4KEQ9 B4ISK9 B4IMC2 B3NKK0 B4NSS8 Q8MSF5 A0A1W4VF31 A0A088AJC6 A0A2A3E8M6 T1D5L0 Q7Z0Q2 B4MJ04 A0A084VZ21 A0A023EEH7 A0A1J1I5D4 A0A0L0C314 B3MBR6 A0A182JZ97 A0A182WC98 A0A2P8YJQ9 B4J9F1 Q1HRK4 A0A182WTU5 Q7Q5K0 A0A182HI42 R4WRC1 A0A182R5J7 A0A1B0GI92 A0A1L8D8Q5 R4G495 A0A182Q8H1 A0A2M3Z1G3 A0A182PET5 A0A2M4ADV5 A0A2M4C4H2 W5JR74 A0A182FEL0 D3PFQ2 A0A2M4A704 A6YPE7 J9KM02 A0A0V0G565 A0A023FB88 A0A336M0C3 A0A0M4ECV7 A0A069DVD2 A0A224Y1Z2 B4LPH6 E9G2F9 A0A195BKP1 A0A1B0B5C1 A0A1B0A3N9 D3TM88 A0A0N8B6T8 A0A0N8A335 A0A0P5TMI1 A0A0P5J892 A0A0N8AHA3 A0A0P4YYM6 A0A0L7RBZ3 D3PHH8 C4N185 A0A1I8N878 A0A1L8EEZ5 A0A1I8PAY8 A0A1D2NAB6
Ontologies
PANTHER
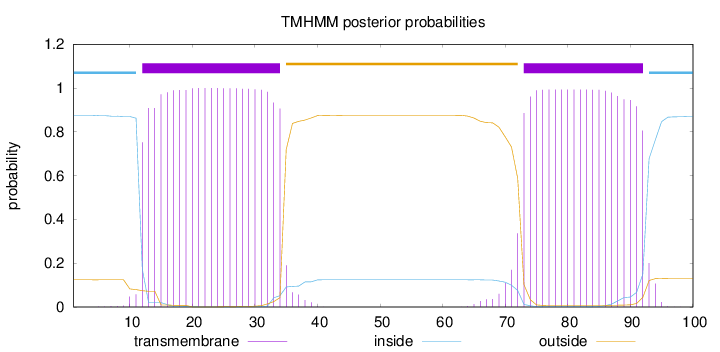
Topology
Subcellular location
Membrane
Length:
100
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.20689
Exp number, first 60 AAs:
22.77887
Total prob of N-in:
0.87583
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 72
TMhelix
73 - 92
inside
93 - 100
Population Genetic Test Statistics
Pi
211.003961
Theta
189.96848
Tajima's D
0.425
CLR
60.476966
CSRT
0.494475276236188
Interpretation
Uncertain
