Gene
KWMTBOMO11571 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004029
Annotation
PREDICTED:_pleckstrin_homology_domain-containing_family_F_member_2_isoform_X2_[Bombyx_mori]
Full name
alpha-1,2-Mannosidase
Location in the cell
Nuclear Reliability : 4.559
Sequence
CDS
ATGGTGGATCGTTTAGTGAATAGTGAAGCAAATGCAAGAAGAATTGCAATGGTGGAGAGCTGCTTCGGCAGCTCGGGACAACCTCTAGCAGAGCAAGGGCGGGTCCTTGTTGGTGAAGGTGTCTTGACAAAAATTTGCAGGAAAAAACCAAAGGCACGACAATTCTTTTTGTTTAATGACATTTTGGTGTATGGCAATATAGTTATTAACAAGAAAAAATACAACAAGCAACATATCATTCCATTAGAGGAGGTTAAGCTGGAGTCACTGAAAGATGAAGGACAATACCGTAATGGATGGTTAATCCGTACTGCATCCAAATCATTTGCTGTGTATGCTGCTACAGCCACAGAAAAAGAAGAATGGATGGCTCATATTGAAAAATGTATAGAAGATTTATTAAGAAAAAGTGGCAAGCAACCTCCTTCAGAGCATGCAGCTGTTTGGGTACCAGACAATGAAGCCTCAATATGTATGCACTGTAAGAAAACACAGTTCACTGTGATAAACAGAAGGCATCATTGCCGCAAGTGCGGATCCGTGGTGTGTGGTCCGTGCTCGTCGAAGCGGTACATCCTGCGCGGCCAGAGCGACAAGCCGCTGCGCGTGTGCCTGCAGTGCTACGACGAGCTCAGCAGGGAACGCGTCCGACCCCCCAACCAGCAGCAGACCTCGGCCAACACCACGCCCGTTAAAACCGATACGGCCGGTTCCGGAGGGGACTCCTCGGGCGACGAAGACAGTGATGACGATGATATCAATGCTGAAGAGAAACACGACGAGCCAAAGTTCTACGGTGACAGCGAAAAGACTGAAGAGACCAACAATCATTCGTCCAAGGAAGCCGCCAAGTAA
Protein
MVDRLVNSEANARRIAMVESCFGSSGQPLAEQGRVLVGEGVLTKICRKKPKARQFFLFNDILVYGNIVINKKKYNKQHIIPLEEVKLESLKDEGQYRNGWLIRTASKSFAVYAATATEKEEWMAHIEKCIEDLLRKSGKQPPSEHAAVWVPDNEASICMHCKKTQFTVINRRHHCRKCGSVVCGPCSSKRYILRGQSDKPLRVCLQCYDELSRERVRPPNQQQTSANTTPVKTDTAGSGGDSSGDEDSDDDDINAEEKHDEPKFYGDSEKTEETNNHSSKEAAK
Summary
Similarity
Belongs to the glycosyl hydrolase 47 family.
Uniprot
H9J3E2
A0A194QXE2
A0A1E1WDT1
A0A194QCX5
A0A2A4K718
A0A1E1WSK9
+ More
A0A2H1WFT7 A0A1E1W5T9 A0A212EX47 A0A1E1VZ17 A0A2J7PU68 A0A224XHG3 A0A023FCR3 A0A069DYD1 A0A1B6LVC2 A0A0V0G813 A0A2J7PU46 A0A0P4VYR5 A0A1B6FNB4 V5GU88 A0A182K8U3 A0A1B6KFC3 A0A1B6CMS0 A0A182J0U6 A0A1Y1LIE2 A0A182VPT7 A0A1B0GJR8 A0A1L8DD50 A0A0A9YB11 A0A2S2QZZ7 A0A158P3Y2 A0A1L8DDK1 A0A182QNU3 A0A1S4H3N6 A6YPS6 A0A232ERB1 J9K8V3 A0A2H8TZR7 A0A023EMW5 A0A139WEK4 A0A182RPK6 A0A088AJ29 A0A1Q3FWR7 K7IS81 A0A2M4BWR9 A0A182NNN0 A0A2M3ZGZ3 A0A2M4AE19 A0A0P4VSQ0 A0A182MFP6 A0A1S4G271 A0A0C9QXW2 W8B9X0 A0A034VVZ8 A0A1Q3FWX8 E9J1Q0 A0A0A1WVB7 A0A2M4AEC7 Q1DH65 A0A1B0A0A5 A0A1Q3F6P7 A0A1B0FAI8 A0A182SZS3 A0A0K8U8R9 A0A1A9ULM8 N6TT54 A0A087TG54 B0X7A7 A0A182YN34 U5EVQ9 Q7PPP7 E9GYI5 T1P9E8 A0A2P6LJA7 A0A182GBU6 A0A0P5G799 A0A0P6HBP9 A0A0P5VVM6 A0A3L8D8F1 A0A3R7ND93 B3MRW7 A0A0K8TQV4 A0A1W4VF51 A0A1I8PEK4 B4MB31 A0A0L0C8R5 W5JV44 A0A151J8Y0 A0A151X0I7 A0A1L8DCG1 A0A151JVD9 A0A195CPY2 A0A293N3D0 V5HGE8 A0A131XC38 A0A0M5J0S0 A0A131YJT5 L7MCU4 A0A224Z686 L7MBL7
A0A2H1WFT7 A0A1E1W5T9 A0A212EX47 A0A1E1VZ17 A0A2J7PU68 A0A224XHG3 A0A023FCR3 A0A069DYD1 A0A1B6LVC2 A0A0V0G813 A0A2J7PU46 A0A0P4VYR5 A0A1B6FNB4 V5GU88 A0A182K8U3 A0A1B6KFC3 A0A1B6CMS0 A0A182J0U6 A0A1Y1LIE2 A0A182VPT7 A0A1B0GJR8 A0A1L8DD50 A0A0A9YB11 A0A2S2QZZ7 A0A158P3Y2 A0A1L8DDK1 A0A182QNU3 A0A1S4H3N6 A6YPS6 A0A232ERB1 J9K8V3 A0A2H8TZR7 A0A023EMW5 A0A139WEK4 A0A182RPK6 A0A088AJ29 A0A1Q3FWR7 K7IS81 A0A2M4BWR9 A0A182NNN0 A0A2M3ZGZ3 A0A2M4AE19 A0A0P4VSQ0 A0A182MFP6 A0A1S4G271 A0A0C9QXW2 W8B9X0 A0A034VVZ8 A0A1Q3FWX8 E9J1Q0 A0A0A1WVB7 A0A2M4AEC7 Q1DH65 A0A1B0A0A5 A0A1Q3F6P7 A0A1B0FAI8 A0A182SZS3 A0A0K8U8R9 A0A1A9ULM8 N6TT54 A0A087TG54 B0X7A7 A0A182YN34 U5EVQ9 Q7PPP7 E9GYI5 T1P9E8 A0A2P6LJA7 A0A182GBU6 A0A0P5G799 A0A0P6HBP9 A0A0P5VVM6 A0A3L8D8F1 A0A3R7ND93 B3MRW7 A0A0K8TQV4 A0A1W4VF51 A0A1I8PEK4 B4MB31 A0A0L0C8R5 W5JV44 A0A151J8Y0 A0A151X0I7 A0A1L8DCG1 A0A151JVD9 A0A195CPY2 A0A293N3D0 V5HGE8 A0A131XC38 A0A0M5J0S0 A0A131YJT5 L7MCU4 A0A224Z686 L7MBL7
EC Number
3.2.1.-
Pubmed
19121390
26354079
22118469
25474469
26334808
27129103
+ More
28004739 25401762 26823975 21347285 12364791 18207082 28648823 24945155 18362917 19820115 20075255 17510324 24495485 25348373 21282665 25830018 23537049 25244985 21292972 25315136 26483478 30249741 17994087 18057021 26369729 26108605 20920257 23761445 25765539 28049606 26830274 25576852 28797301
28004739 25401762 26823975 21347285 12364791 18207082 28648823 24945155 18362917 19820115 20075255 17510324 24495485 25348373 21282665 25830018 23537049 25244985 21292972 25315136 26483478 30249741 17994087 18057021 26369729 26108605 20920257 23761445 25765539 28049606 26830274 25576852 28797301
EMBL
BABH01021510
BABH01021511
KQ461073
KPJ09625.1
GDQN01005939
JAT85115.1
+ More
KQ459185 KPJ03317.1 NWSH01000111 PCG79450.1 GDQN01001100 JAT89954.1 ODYU01008049 SOQ51324.1 GDQN01008730 JAT82324.1 AGBW02011827 OWR46072.1 GDQN01011071 JAT79983.1 NEVH01021208 PNF19864.1 GFTR01004489 JAW11937.1 GBBI01000089 JAC18623.1 GBGD01002470 JAC86419.1 GEBQ01012354 JAT27623.1 GECL01001966 JAP04158.1 PNF19865.1 GDKW01000837 JAI55758.1 GECZ01018072 JAS51697.1 GALX01003239 JAB65227.1 GEBQ01030093 JAT09884.1 GEDC01022529 JAS14769.1 GEZM01054776 JAV73403.1 AJWK01022156 AJWK01022157 AJWK01022158 AJWK01022159 AJWK01022160 GFDF01009691 JAV04393.1 GBHO01014240 GBRD01012558 GBRD01012556 GBRD01012555 GBRD01012551 GBRD01003291 GDHC01020392 JAG29364.1 JAG53266.1 JAP98236.1 GGMS01014128 MBY83331.1 ADTU01008610 GFDF01009690 JAV04394.1 AXCN02001878 AAAB01008943 EF639087 ABR27972.1 NNAY01002629 OXU20903.1 ABLF02034425 GFXV01007988 MBW19793.1 GAPW01003398 JAC10200.1 KQ971354 KYB26366.1 GFDL01002995 JAV32050.1 GGFJ01008376 MBW57517.1 GGFM01007053 MBW27804.1 GGFK01005710 MBW39031.1 GDRN01100469 JAI58567.1 AXCM01002503 GBYB01005527 GBYB01005528 JAG75294.1 JAG75295.1 GAMC01012715 GAMC01012713 GAMC01012712 GAMC01012711 JAB93843.1 GAKP01012692 GAKP01012691 GAKP01012690 JAC46262.1 GFDL01002997 JAV32048.1 GL767674 EFZ13287.1 GBXI01011273 GBXI01008704 GBXI01001116 GBXI01000665 JAD03019.1 JAD05588.1 JAD13176.1 JAD13627.1 GGFK01005820 MBW39141.1 CH899796 EAT32501.1 GFDL01011804 JAV23241.1 CCAG010006942 GDHF01033345 GDHF01029458 GDHF01029343 GDHF01025347 GDHF01021440 GDHF01001777 JAI18969.1 JAI22856.1 JAI22971.1 JAI26967.1 JAI30874.1 JAI50537.1 APGK01019947 KB740150 KB632173 ENN81243.1 ERL89568.1 KK115064 KFM64093.1 DS232445 EDS41860.1 GANO01001766 JAB58105.1 EAA10045.5 GL732575 EFX75476.1 KA644785 AFP59414.1 MWRG01000020 PRD38671.1 JXUM01053341 JXUM01053342 JXUM01053343 KQ561772 KXJ77589.1 GDIQ01246262 JAK05463.1 GDIP01237000 GDIQ01022487 LRGB01002864 JAI86401.1 JAN72250.1 KZS05772.1 GDIP01094562 JAM09153.1 QOIP01000011 RLU16392.1 QCYY01000597 ROT84067.1 CH902622 EDV34522.1 KPU75184.1 GDAI01001075 JAI16528.1 CH940655 EDW66440.1 JRES01000755 KNC28666.1 ADMH02000041 ETN68016.1 KQ979480 KYN21279.1 KQ982612 KYQ53866.1 GFDF01009942 JAV04142.1 KQ981701 KYN37493.1 KQ977513 KYN02169.1 GFWV01022559 MAA47286.1 GANP01007859 JAB76609.1 GEFH01004394 JAP64187.1 CP012528 ALC48298.1 GEDV01010386 JAP78171.1 GACK01003402 JAA61632.1 GFPF01010644 MAA21790.1 GACK01003403 JAA61631.1
KQ459185 KPJ03317.1 NWSH01000111 PCG79450.1 GDQN01001100 JAT89954.1 ODYU01008049 SOQ51324.1 GDQN01008730 JAT82324.1 AGBW02011827 OWR46072.1 GDQN01011071 JAT79983.1 NEVH01021208 PNF19864.1 GFTR01004489 JAW11937.1 GBBI01000089 JAC18623.1 GBGD01002470 JAC86419.1 GEBQ01012354 JAT27623.1 GECL01001966 JAP04158.1 PNF19865.1 GDKW01000837 JAI55758.1 GECZ01018072 JAS51697.1 GALX01003239 JAB65227.1 GEBQ01030093 JAT09884.1 GEDC01022529 JAS14769.1 GEZM01054776 JAV73403.1 AJWK01022156 AJWK01022157 AJWK01022158 AJWK01022159 AJWK01022160 GFDF01009691 JAV04393.1 GBHO01014240 GBRD01012558 GBRD01012556 GBRD01012555 GBRD01012551 GBRD01003291 GDHC01020392 JAG29364.1 JAG53266.1 JAP98236.1 GGMS01014128 MBY83331.1 ADTU01008610 GFDF01009690 JAV04394.1 AXCN02001878 AAAB01008943 EF639087 ABR27972.1 NNAY01002629 OXU20903.1 ABLF02034425 GFXV01007988 MBW19793.1 GAPW01003398 JAC10200.1 KQ971354 KYB26366.1 GFDL01002995 JAV32050.1 GGFJ01008376 MBW57517.1 GGFM01007053 MBW27804.1 GGFK01005710 MBW39031.1 GDRN01100469 JAI58567.1 AXCM01002503 GBYB01005527 GBYB01005528 JAG75294.1 JAG75295.1 GAMC01012715 GAMC01012713 GAMC01012712 GAMC01012711 JAB93843.1 GAKP01012692 GAKP01012691 GAKP01012690 JAC46262.1 GFDL01002997 JAV32048.1 GL767674 EFZ13287.1 GBXI01011273 GBXI01008704 GBXI01001116 GBXI01000665 JAD03019.1 JAD05588.1 JAD13176.1 JAD13627.1 GGFK01005820 MBW39141.1 CH899796 EAT32501.1 GFDL01011804 JAV23241.1 CCAG010006942 GDHF01033345 GDHF01029458 GDHF01029343 GDHF01025347 GDHF01021440 GDHF01001777 JAI18969.1 JAI22856.1 JAI22971.1 JAI26967.1 JAI30874.1 JAI50537.1 APGK01019947 KB740150 KB632173 ENN81243.1 ERL89568.1 KK115064 KFM64093.1 DS232445 EDS41860.1 GANO01001766 JAB58105.1 EAA10045.5 GL732575 EFX75476.1 KA644785 AFP59414.1 MWRG01000020 PRD38671.1 JXUM01053341 JXUM01053342 JXUM01053343 KQ561772 KXJ77589.1 GDIQ01246262 JAK05463.1 GDIP01237000 GDIQ01022487 LRGB01002864 JAI86401.1 JAN72250.1 KZS05772.1 GDIP01094562 JAM09153.1 QOIP01000011 RLU16392.1 QCYY01000597 ROT84067.1 CH902622 EDV34522.1 KPU75184.1 GDAI01001075 JAI16528.1 CH940655 EDW66440.1 JRES01000755 KNC28666.1 ADMH02000041 ETN68016.1 KQ979480 KYN21279.1 KQ982612 KYQ53866.1 GFDF01009942 JAV04142.1 KQ981701 KYN37493.1 KQ977513 KYN02169.1 GFWV01022559 MAA47286.1 GANP01007859 JAB76609.1 GEFH01004394 JAP64187.1 CP012528 ALC48298.1 GEDV01010386 JAP78171.1 GACK01003402 JAA61632.1 GFPF01010644 MAA21790.1 GACK01003403 JAA61631.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000235965
+ More
UP000075881 UP000075880 UP000075920 UP000092461 UP000005205 UP000075886 UP000215335 UP000007819 UP000007266 UP000075900 UP000005203 UP000002358 UP000075884 UP000075883 UP000008820 UP000092445 UP000092444 UP000075901 UP000078200 UP000019118 UP000030742 UP000054359 UP000002320 UP000076408 UP000007062 UP000000305 UP000095301 UP000069940 UP000249989 UP000076858 UP000279307 UP000283509 UP000007801 UP000192221 UP000095300 UP000008792 UP000037069 UP000000673 UP000078492 UP000075809 UP000078541 UP000078542 UP000092553
UP000075881 UP000075880 UP000075920 UP000092461 UP000005205 UP000075886 UP000215335 UP000007819 UP000007266 UP000075900 UP000005203 UP000002358 UP000075884 UP000075883 UP000008820 UP000092445 UP000092444 UP000075901 UP000078200 UP000019118 UP000030742 UP000054359 UP000002320 UP000076408 UP000007062 UP000000305 UP000095301 UP000069940 UP000249989 UP000076858 UP000279307 UP000283509 UP000007801 UP000192221 UP000095300 UP000008792 UP000037069 UP000000673 UP000078492 UP000075809 UP000078541 UP000078542 UP000092553
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J3E2
A0A194QXE2
A0A1E1WDT1
A0A194QCX5
A0A2A4K718
A0A1E1WSK9
+ More
A0A2H1WFT7 A0A1E1W5T9 A0A212EX47 A0A1E1VZ17 A0A2J7PU68 A0A224XHG3 A0A023FCR3 A0A069DYD1 A0A1B6LVC2 A0A0V0G813 A0A2J7PU46 A0A0P4VYR5 A0A1B6FNB4 V5GU88 A0A182K8U3 A0A1B6KFC3 A0A1B6CMS0 A0A182J0U6 A0A1Y1LIE2 A0A182VPT7 A0A1B0GJR8 A0A1L8DD50 A0A0A9YB11 A0A2S2QZZ7 A0A158P3Y2 A0A1L8DDK1 A0A182QNU3 A0A1S4H3N6 A6YPS6 A0A232ERB1 J9K8V3 A0A2H8TZR7 A0A023EMW5 A0A139WEK4 A0A182RPK6 A0A088AJ29 A0A1Q3FWR7 K7IS81 A0A2M4BWR9 A0A182NNN0 A0A2M3ZGZ3 A0A2M4AE19 A0A0P4VSQ0 A0A182MFP6 A0A1S4G271 A0A0C9QXW2 W8B9X0 A0A034VVZ8 A0A1Q3FWX8 E9J1Q0 A0A0A1WVB7 A0A2M4AEC7 Q1DH65 A0A1B0A0A5 A0A1Q3F6P7 A0A1B0FAI8 A0A182SZS3 A0A0K8U8R9 A0A1A9ULM8 N6TT54 A0A087TG54 B0X7A7 A0A182YN34 U5EVQ9 Q7PPP7 E9GYI5 T1P9E8 A0A2P6LJA7 A0A182GBU6 A0A0P5G799 A0A0P6HBP9 A0A0P5VVM6 A0A3L8D8F1 A0A3R7ND93 B3MRW7 A0A0K8TQV4 A0A1W4VF51 A0A1I8PEK4 B4MB31 A0A0L0C8R5 W5JV44 A0A151J8Y0 A0A151X0I7 A0A1L8DCG1 A0A151JVD9 A0A195CPY2 A0A293N3D0 V5HGE8 A0A131XC38 A0A0M5J0S0 A0A131YJT5 L7MCU4 A0A224Z686 L7MBL7
A0A2H1WFT7 A0A1E1W5T9 A0A212EX47 A0A1E1VZ17 A0A2J7PU68 A0A224XHG3 A0A023FCR3 A0A069DYD1 A0A1B6LVC2 A0A0V0G813 A0A2J7PU46 A0A0P4VYR5 A0A1B6FNB4 V5GU88 A0A182K8U3 A0A1B6KFC3 A0A1B6CMS0 A0A182J0U6 A0A1Y1LIE2 A0A182VPT7 A0A1B0GJR8 A0A1L8DD50 A0A0A9YB11 A0A2S2QZZ7 A0A158P3Y2 A0A1L8DDK1 A0A182QNU3 A0A1S4H3N6 A6YPS6 A0A232ERB1 J9K8V3 A0A2H8TZR7 A0A023EMW5 A0A139WEK4 A0A182RPK6 A0A088AJ29 A0A1Q3FWR7 K7IS81 A0A2M4BWR9 A0A182NNN0 A0A2M3ZGZ3 A0A2M4AE19 A0A0P4VSQ0 A0A182MFP6 A0A1S4G271 A0A0C9QXW2 W8B9X0 A0A034VVZ8 A0A1Q3FWX8 E9J1Q0 A0A0A1WVB7 A0A2M4AEC7 Q1DH65 A0A1B0A0A5 A0A1Q3F6P7 A0A1B0FAI8 A0A182SZS3 A0A0K8U8R9 A0A1A9ULM8 N6TT54 A0A087TG54 B0X7A7 A0A182YN34 U5EVQ9 Q7PPP7 E9GYI5 T1P9E8 A0A2P6LJA7 A0A182GBU6 A0A0P5G799 A0A0P6HBP9 A0A0P5VVM6 A0A3L8D8F1 A0A3R7ND93 B3MRW7 A0A0K8TQV4 A0A1W4VF51 A0A1I8PEK4 B4MB31 A0A0L0C8R5 W5JV44 A0A151J8Y0 A0A151X0I7 A0A1L8DCG1 A0A151JVD9 A0A195CPY2 A0A293N3D0 V5HGE8 A0A131XC38 A0A0M5J0S0 A0A131YJT5 L7MCU4 A0A224Z686 L7MBL7
PDB
3MPX
E-value=1.4941e-15,
Score=200
Ontologies
GO
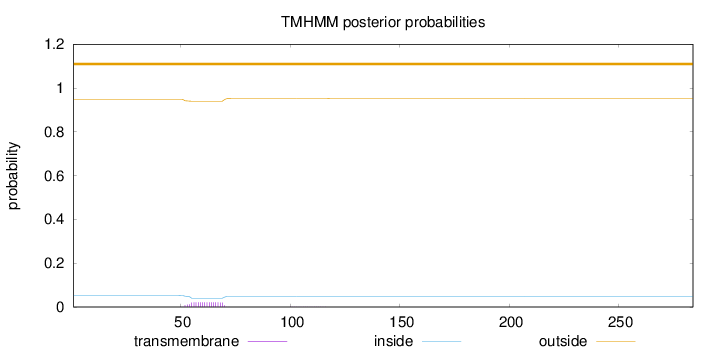
Topology
Length:
284
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.35356
Exp number, first 60 AAs:
0.15685
Total prob of N-in:
0.05225
outside
1 - 284
Population Genetic Test Statistics
Pi
218.012189
Theta
165.71052
Tajima's D
0.537857
CLR
0.033302
CSRT
0.52262386880656
Interpretation
Uncertain
