Gene
KWMTBOMO11568
Pre Gene Modal
BGIBMGA004089
Annotation
PREDICTED:_tyrosine-protein_kinase_Src42A-like_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.336
Sequence
CDS
ATGGTGTCTGAGGGGGCTTTGCCCGTAGTGTGGTGTATACGCGATAATCAACTTGTGCCGCATCAGGTATCCGGTGTAAAATTACAAGGAGCTGTTACCGGAGCGCATATAGCTGCAGTTACTGATTTCTTGCGAATGGTCGCCGGTACTCCGATAGTTCTACCACATACTTCTGTTACAGAAGACCTTGAAAATGATATCAATCAACAGAAAGAAAATTCTACAAGAAGACGTAAAATGTCTATGCAAGTGGAAGTGCCATGTGTATTCCTCTCTGAAAGTGCGCTACAGAAGGCGAAAAGTCGTAATAATTCTCGTGAGGAAAAGGTAACAACATTTGAAAATTTGTCAGCGGTAGCTAAAAAACTTTTAGAATTAGAAAATGCACCAATTCTATCGAATGGTTTAGAATATATTGACACGCAGGAGCCTTTGGAGGATCTTTGTGTAGAAGTAAATACTATAAAACAAAAATCTATAGATACTAAAACCACAAATCACGACATCATTTCACAAAGAATAACTAGAATAATAGATCTGAATGATGGGTTAGAAACCAACAATATGGGTCACATTGAGTGTGAACAAGAATCACCCCAAAGAATCACGAGAATCATTGAATTTGAAAATGATAAAACTACAAAATTCATAGAAAATGAAACACCGCAAAGAATTACTAGAATTATAGAACTTAACGAAGATAATTCTTTTTCAACAATCACAGAAAATCATGAAACTGATTCAAGTCAAAAGTTAAGTCGAATAGATGAATTAGAGAAAGAAAATGTTTTTCCTAGCATGACAAAAATTATCGAAATGGAGAATGTTATCTCAAATGAACGAAGAATCTCTTTAACAAACGAACGTGAAACTGGTGGTGGCAAAATTGAGCCTTCTTGCGATGAAGTTATTGTAAGGTCTAGGTATATTCATAAAAGTGCCCCTAGGTTTAGTCTTGATTCCTCTTGCTCCGAAAACTCTTACGTTAATATTGACTGCCTGAAGAGTTTGCAGGATGTTAAAAAAGAGGACATTGTGGATGTAAGCGCGAACACTGAAACTTCACCCACTAGTAGATGTATTCGTTCTCGCTTTCCACGAAAAAAAACTTCTGTCTGTTCAATTGGTTCATCTGATTGCGATGATGAAGTCGATGTGATATTCAAATCTAAACCTAAATCTAGTCAGTGGGTAAGTTTGGATTGGATTCCGCCTCCGCCAAAAGAAGATCCAGTGATTGCACAAGTCTGTGATAAAAATGATTTCATTACGAAATGGATAGCTGATCAAAATTCGGTGATCGTTGCCGACGAAAGAAGAAAAAGTCTTCCACCAAAATCTAATGAAATTTATTTACAAAACATGAGGAGGTTTAGTGATGGTCTACAAATATGTAAAGAAGATGCAGCCAGTGATTCGCCCGCTACAGTGAAATGGAAGTGGCAAGATCTCGTCAAACGCCACATCCAGTTACTTAAATCTGACAAAGGATTCCGACGACAAAGAGGCAGCTGGATGCGAACTGCAAGGCGCATGTCTGGAACTAGCGATAACAATAAGAACTTGGACACACAGAATTTGTATGCCCACATGAAATTAAATAATATGCTATGGTACTTCCGCAAAATAAAACGGATCGAAGCGGAAAAGAAACTGTTACTACCCGAAAACGAGCATGGCGCTTTCCTTATCCGTGATTCTGAGAGCCGCCACAATGACTTCTCACTATCAGTACGAGACGGTGACACAGTGAAACACTATCGCATCCGGCAACTGGACGAGGGAGGGTTTTTCATAGCCCGACGCACAACCTTCCGCACACTTCAAGAACTCGTCGAGCACTACAGCAAGGACGCAGACGGACTATGCGTCGCCCTCAATAAACCTTGCGTTCAGATCGAGAAACCGGTGACAGAAGGCCTGTCTCACAGAACACGTGATCAATGGGAAATCGATCGTGCATCATTGAAGTTTGTTCGGAAACTCGGCCACGGTCAGTTCGGAGAAGTATGGGAAGGATTGTGGAACAACACGACTCCGGTCGCGGTGAAAACTCTCAAATCCGGCACAATGGACCCAAAGGATTTCCTCGCTGAAGCGCAAATCATGAAGAAGCTGAGGCACAACAAACTAATCCAGCTATACGCTGTGTGTACTCTTGAAGAGCCTATCTACATAATCACCGAACTCATGAAACACGGTTCTTTATTGGATTATTTACAAGGTAAAGGTCGCGGTCTGAAACTGCAGCAGCTGATCGACATGGCAGCCCAGATAGCGGCCGGCATGGCGTATCTGGAATCGCAGAACTACATCCACCGAGACCTCGCGGCCAGGAACGTCCTCGTGGGCGACGGTAACATCGTCAAGATCGCCGACTTCGGACTGGCCAGACTGATCAAAGAAGACGAATACGAAGCTCGCGTGGGTGCAAGGTTCCCTATAAAATGGACAGCACCCGAAGCGGCGAACTACAGCAAGTTTTCAATCAAATCTGACGTCTGGTCGTTTGGAATCCTGCTTACTGAGCTCGTCACGTACGGCCGGACCCCTTACCCAGGCATGACCAATGCTGAAGTACTACACCAAGTGGAACACGGGTACCGCATGCCGTGCCCGCAGAATTGTCTGCCGGCCCTCTACGAAATCATGCTGGAGTGTTGGCACAAGGATCCCCTCAAACGGCCCACCTTCGAGACCCTGCAGTGGAAGCTCGAAGACTTCTTCACCATGGACAACTCGGAGTACAAGGAGGCGTCCACCTACTGA
Protein
MVSEGALPVVWCIRDNQLVPHQVSGVKLQGAVTGAHIAAVTDFLRMVAGTPIVLPHTSVTEDLENDINQQKENSTRRRKMSMQVEVPCVFLSESALQKAKSRNNSREEKVTTFENLSAVAKKLLELENAPILSNGLEYIDTQEPLEDLCVEVNTIKQKSIDTKTTNHDIISQRITRIIDLNDGLETNNMGHIECEQESPQRITRIIEFENDKTTKFIENETPQRITRIIELNEDNSFSTITENHETDSSQKLSRIDELEKENVFPSMTKIIEMENVISNERRISLTNERETGGGKIEPSCDEVIVRSRYIHKSAPRFSLDSSCSENSYVNIDCLKSLQDVKKEDIVDVSANTETSPTSRCIRSRFPRKKTSVCSIGSSDCDDEVDVIFKSKPKSSQWVSLDWIPPPPKEDPVIAQVCDKNDFITKWIADQNSVIVADERRKSLPPKSNEIYLQNMRRFSDGLQICKEDAASDSPATVKWKWQDLVKRHIQLLKSDKGFRRQRGSWMRTARRMSGTSDNNKNLDTQNLYAHMKLNNMLWYFRKIKRIEAEKKLLLPENEHGAFLIRDSESRHNDFSLSVRDGDTVKHYRIRQLDEGGFFIARRTTFRTLQELVEHYSKDADGLCVALNKPCVQIEKPVTEGLSHRTRDQWEIDRASLKFVRKLGHGQFGEVWEGLWNNTTPVAVKTLKSGTMDPKDFLAEAQIMKKLRHNKLIQLYAVCTLEEPIYIITELMKHGSLLDYLQGKGRGLKLQQLIDMAAQIAAGMAYLESQNYIHRDLAARNVLVGDGNIVKIADFGLARLIKEDEYEARVGARFPIKWTAPEAANYSKFSIKSDVWSFGILLTELVTYGRTPYPGMTNAEVLHQVEHGYRMPCPQNCLPALYEIMLECWHKDPLKRPTFETLQWKLEDFFTMDNSEYKEASTY
Summary
Uniprot
EMBL
Pfam
PF00017 SH2
SUPFAM
SSF55550
SSF55550
Gene 3D
ProteinModelPortal
PDB
2PTK
E-value=1.62077e-135,
Score=1240
Ontologies
GO
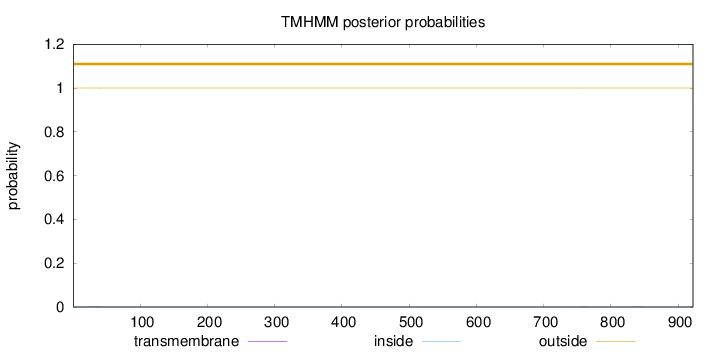
Topology
Length:
922
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0105
Exp number, first 60 AAs:
0.00454
Total prob of N-in:
0.00022
outside
1 - 922
Population Genetic Test Statistics
Pi
164.331234
Theta
158.1207
Tajima's D
0.045568
CLR
0.699348
CSRT
0.375131243437828
Interpretation
Possibly Positive selection
