Gene
KWMTBOMO11566
Pre Gene Modal
BGIBMGA004032
Annotation
neuropeptide_receptor_A20_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.726
Sequence
CDS
ATGCTGTCGGAAAACGAAACCCGAAGTAGTTACGACACTCAGACAAATCTTTCCCACATCTCCGGCAATTTGTTTGAAGATGCCAGTATAAATTCCCTATTGAACCACGGCGGGAATTACACTACAAACATCGGACAAGTGCTCAAAGTGCTCGAAGATTGGAGAAACAGATTCAATATAACCGTTGTTAAAGAATGCGATGGAACAGAATACTGCGCTGGGGAGTTCCGAGATTTGATAATAGCATACAACAGTATTCACGGCTACGTCAGTTTGTTGGTCTGTCTTTTTGGTAGTCTCGCCAACGCTTTTAATGTAGCCGTGCTGACGAGGCGAGATCTCGCAGTGGCTCCCATAAATCGACTGCTAAAATGGCTTGCAGTAGCTGACGTCTTCGTCATGATTGAGTATGTTCCATTCGCTATCTACAGGTATTTGTTGTTACCCGGTCAAGAAGACCGTCCATACTCCTGGGCTGCATATATGCTGTTTCATATGCACTTCACGCAGATATTCCATACAGCGTCAATACTGTTAACCTTGTCTCTGGCAGTTTGGCGATATCTTGCAATCAAATATCCGGCTCACAGTCCAATATTATGCACAGATAGGCGATGCACAGTGGCCATAATGTTGAGTTTTGTGCTTCCCTCTATACTCTGCATACCATCGTACTTTGTCTTCACAATCCATAAAGACTTCTCATACGACAGGAACGCCAACGTTTTCTCGAAAGTTTATTTCGTAGATTCCGATTTCGACGGATACCTCTATCATATTAATTTCTGGATCGACAAAACGGTCTAA
Protein
MLSENETRSSYDTQTNLSHISGNLFEDASINSLLNHGGNYTTNIGQVLKVLEDWRNRFNITVVKECDGTEYCAGEFRDLIIAYNSIHGYVSLLVCLFGSLANAFNVAVLTRRDLAVAPINRLLKWLAVADVFVMIEYVPFAIYRYLLLPGQEDRPYSWAAYMLFHMHFTQIFHTASILLTLSLAVWRYLAIKYPAHSPILCTDRRCTVAIMLSFVLPSILCIPSYFVFTIHKDFSYDRNANVFSKVYFVDSDFDGYLYHINFWIDKTV
Summary
Uniprot
B3XXN3
A0A2A4JF21
A0A2W1BS69
H9J3E5
A0A194QCR1
A0A194QVS4
+ More
A0A212EX53 A0A2H1VIC9 A0A0N0P9E7 A0A0S1YD84 A0A2W1BU96 A0A194QJU0 B3XXL5 A0A2W1BS81 A0A194QVY6 A0A0S1YD68 A0A212FLW4 B3XXM1 A0A194QK98 A0A0S1YD70 A0A3S2LDK4 A0A182FEC0 A0A084W9M0 A0A182PTL8 A0A182HXV4 A0A182WS57 Q7PPU4 A0A182GLH9 A0A1B0GIH6 E2A775 A0A182IL33 E0VMH3 A0A182YBX8 A0A182NV16 A0A182K6S2 A0A1A9W2D3 A0A182QC09 A0A0C9R9E7 A0A182RYZ1 A0A0L0BRH5 A0A182WMI1 A0A0Q9WB25 A0A1A9Z260 B0WHK7 A0A1A9V040 A0A1A9Y7I5 A0A1B0GFF7 A0A1B0BG75 A0A0C9R2H4 Q7JVS8 B4P7L2 B4HN36 B4ME18 A0A1W4UI32 A0A182RY01 B4QB29 B4KQ21 A0A034WSM1 A0A0K8U7M5 F8J1K8 A0A088A650 B4J7X1 A0A2A3EJ03 B3NSN7 A0A182LHV6 B4H6C2 Q28WS2 B3MEQ2 A0A1I8PDP1 A0A0K8WGQ5 A0A1I8MJD1 B4MK79 A0A1S4FJS5 Q16YF4 A0A1B6I7F6 A0A0R3NPY6 U3U8X9 D6WTY0 A0A0J7KTA3 K7J443 A0A336LPL8 E2BHK2 A0A0M5IZK7 A0A1Y1MWW1 A0A139WAQ6 D6X512 A0A3B0J3V8 A0A1B6ECD2 T1J538
A0A212EX53 A0A2H1VIC9 A0A0N0P9E7 A0A0S1YD84 A0A2W1BU96 A0A194QJU0 B3XXL5 A0A2W1BS81 A0A194QVY6 A0A0S1YD68 A0A212FLW4 B3XXM1 A0A194QK98 A0A0S1YD70 A0A3S2LDK4 A0A182FEC0 A0A084W9M0 A0A182PTL8 A0A182HXV4 A0A182WS57 Q7PPU4 A0A182GLH9 A0A1B0GIH6 E2A775 A0A182IL33 E0VMH3 A0A182YBX8 A0A182NV16 A0A182K6S2 A0A1A9W2D3 A0A182QC09 A0A0C9R9E7 A0A182RYZ1 A0A0L0BRH5 A0A182WMI1 A0A0Q9WB25 A0A1A9Z260 B0WHK7 A0A1A9V040 A0A1A9Y7I5 A0A1B0GFF7 A0A1B0BG75 A0A0C9R2H4 Q7JVS8 B4P7L2 B4HN36 B4ME18 A0A1W4UI32 A0A182RY01 B4QB29 B4KQ21 A0A034WSM1 A0A0K8U7M5 F8J1K8 A0A088A650 B4J7X1 A0A2A3EJ03 B3NSN7 A0A182LHV6 B4H6C2 Q28WS2 B3MEQ2 A0A1I8PDP1 A0A0K8WGQ5 A0A1I8MJD1 B4MK79 A0A1S4FJS5 Q16YF4 A0A1B6I7F6 A0A0R3NPY6 U3U8X9 D6WTY0 A0A0J7KTA3 K7J443 A0A336LPL8 E2BHK2 A0A0M5IZK7 A0A1Y1MWW1 A0A139WAQ6 D6X512 A0A3B0J3V8 A0A1B6ECD2 T1J538
Pubmed
18725956
28756777
19121390
26354079
22118469
24438588
+ More
12364791 14747013 17210077 26483478 20798317 20566863 25244985 26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 22936249 25348373 20966253 15632085 25315136 17510324 23185243 23932938 18054377 18025266 18316733 20068045 21843505 23604020 20075255 28004739 18362917 19820115
12364791 14747013 17210077 26483478 20798317 20566863 25244985 26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 22936249 25348373 20966253 15632085 25315136 17510324 23185243 23932938 18054377 18025266 18316733 20068045 21843505 23604020 20075255 28004739 18362917 19820115
EMBL
AB330441
BAG68419.1
NWSH01001613
PCG70685.1
KZ149917
PZC77922.1
+ More
BABH01021493 KQ459185 KPJ03328.1 KQ461073 KPJ09633.1 AGBW02011827 OWR46065.1 ODYU01002477 SOQ40014.1 KQ459476 KPJ00089.1 KT031018 RSAL01000235 ALM88316.1 RVE43771.1 PZC77921.1 KQ458575 KPJ05797.1 AB330423 BAG68401.1 PZC77918.1 KPJ09637.1 KT031000 ALM88298.1 AGBW02007701 OWR54690.1 AB330429 BAG68407.1 KPJ05799.1 KT031006 ALM88304.1 RSAL01000034 RVE51426.1 ATLV01021836 ATLV01021837 ATLV01021838 KE525324 KFB46914.1 APCN01005356 AY856701 AAAB01008905 AAX98671.1 EAA09746.4 JXUM01072193 JXUM01072194 JXUM01072195 JXUM01072196 KQ562707 KXJ75297.1 AJWK01014143 GL437267 EFN70731.1 DS235308 EEB14579.1 AXCN02001008 GBYB01013034 JAG82801.1 JRES01001578 KNC21819.1 CH940662 KRF78484.1 DS231938 EDS27856.1 CCAG010016793 JXJN01013792 GBYB01002190 JAG71957.1 AE013599 AY099098 AY118700 BT081415 AAF58717.1 AAM28948.1 AAM50560.1 ACO51546.1 CM000158 EDW90047.1 KRJ98877.1 CH480816 EDW47340.1 EDW58783.1 KRF78481.1 KRF78482.1 KRF78483.1 KRF78485.1 CM000362 CM002911 EDX06562.1 KMY92906.1 CH933808 EDW08123.1 KRG03939.1 KRG03940.1 KRG03941.1 GAKP01000376 JAC58576.1 GDHF01029738 JAI22576.1 EU785051 ACI90288.1 CH916367 EDW01177.1 KZ288229 PBC31700.1 CH954179 EDV56539.1 CH479213 EDW33346.1 CM000071 EAL26595.1 CH902619 EDV35516.1 GDHF01002090 JAI50224.1 CH963846 EDW72518.1 CH477517 EAT39649.2 GECU01024873 JAS82833.1 KRT03200.1 KRT03201.1 AB817285 BAO01052.1 BK006095 DAA64476.1 LBMM01003344 KMQ93627.1 AAZX01000986 UFQT01000102 SSX20012.1 GL448301 EFN84855.1 CP012524 ALC41748.1 GEZM01018629 JAV90113.1 KQ971381 KYB24979.1 BK006093 DAA64474.1 OUUW01000001 SPP73982.1 GEDC01012998 GEDC01008232 GEDC01001774 GEDC01000521 JAS24300.1 JAS29066.1 JAS35524.1 JAS36777.1 JH431850
BABH01021493 KQ459185 KPJ03328.1 KQ461073 KPJ09633.1 AGBW02011827 OWR46065.1 ODYU01002477 SOQ40014.1 KQ459476 KPJ00089.1 KT031018 RSAL01000235 ALM88316.1 RVE43771.1 PZC77921.1 KQ458575 KPJ05797.1 AB330423 BAG68401.1 PZC77918.1 KPJ09637.1 KT031000 ALM88298.1 AGBW02007701 OWR54690.1 AB330429 BAG68407.1 KPJ05799.1 KT031006 ALM88304.1 RSAL01000034 RVE51426.1 ATLV01021836 ATLV01021837 ATLV01021838 KE525324 KFB46914.1 APCN01005356 AY856701 AAAB01008905 AAX98671.1 EAA09746.4 JXUM01072193 JXUM01072194 JXUM01072195 JXUM01072196 KQ562707 KXJ75297.1 AJWK01014143 GL437267 EFN70731.1 DS235308 EEB14579.1 AXCN02001008 GBYB01013034 JAG82801.1 JRES01001578 KNC21819.1 CH940662 KRF78484.1 DS231938 EDS27856.1 CCAG010016793 JXJN01013792 GBYB01002190 JAG71957.1 AE013599 AY099098 AY118700 BT081415 AAF58717.1 AAM28948.1 AAM50560.1 ACO51546.1 CM000158 EDW90047.1 KRJ98877.1 CH480816 EDW47340.1 EDW58783.1 KRF78481.1 KRF78482.1 KRF78483.1 KRF78485.1 CM000362 CM002911 EDX06562.1 KMY92906.1 CH933808 EDW08123.1 KRG03939.1 KRG03940.1 KRG03941.1 GAKP01000376 JAC58576.1 GDHF01029738 JAI22576.1 EU785051 ACI90288.1 CH916367 EDW01177.1 KZ288229 PBC31700.1 CH954179 EDV56539.1 CH479213 EDW33346.1 CM000071 EAL26595.1 CH902619 EDV35516.1 GDHF01002090 JAI50224.1 CH963846 EDW72518.1 CH477517 EAT39649.2 GECU01024873 JAS82833.1 KRT03200.1 KRT03201.1 AB817285 BAO01052.1 BK006095 DAA64476.1 LBMM01003344 KMQ93627.1 AAZX01000986 UFQT01000102 SSX20012.1 GL448301 EFN84855.1 CP012524 ALC41748.1 GEZM01018629 JAV90113.1 KQ971381 KYB24979.1 BK006093 DAA64474.1 OUUW01000001 SPP73982.1 GEDC01012998 GEDC01008232 GEDC01001774 GEDC01000521 JAS24300.1 JAS29066.1 JAS35524.1 JAS36777.1 JH431850
Proteomes
UP000218220
UP000005204
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000069272 UP000030765 UP000075885 UP000075840 UP000076407 UP000007062 UP000069940 UP000249989 UP000092461 UP000000311 UP000075880 UP000009046 UP000076408 UP000075884 UP000075881 UP000091820 UP000075886 UP000075900 UP000037069 UP000075920 UP000008792 UP000092445 UP000002320 UP000078200 UP000092443 UP000092444 UP000092460 UP000000803 UP000002282 UP000001292 UP000192221 UP000000304 UP000009192 UP000005203 UP000001070 UP000242457 UP000008711 UP000075882 UP000008744 UP000001819 UP000007801 UP000095300 UP000095301 UP000007798 UP000008820 UP000036403 UP000002358 UP000008237 UP000092553 UP000007266 UP000268350
UP000069272 UP000030765 UP000075885 UP000075840 UP000076407 UP000007062 UP000069940 UP000249989 UP000092461 UP000000311 UP000075880 UP000009046 UP000076408 UP000075884 UP000075881 UP000091820 UP000075886 UP000075900 UP000037069 UP000075920 UP000008792 UP000092445 UP000002320 UP000078200 UP000092443 UP000092444 UP000092460 UP000000803 UP000002282 UP000001292 UP000192221 UP000000304 UP000009192 UP000005203 UP000001070 UP000242457 UP000008711 UP000075882 UP000008744 UP000001819 UP000007801 UP000095300 UP000095301 UP000007798 UP000008820 UP000036403 UP000002358 UP000008237 UP000092553 UP000007266 UP000268350
Interpro
ProteinModelPortal
B3XXN3
A0A2A4JF21
A0A2W1BS69
H9J3E5
A0A194QCR1
A0A194QVS4
+ More
A0A212EX53 A0A2H1VIC9 A0A0N0P9E7 A0A0S1YD84 A0A2W1BU96 A0A194QJU0 B3XXL5 A0A2W1BS81 A0A194QVY6 A0A0S1YD68 A0A212FLW4 B3XXM1 A0A194QK98 A0A0S1YD70 A0A3S2LDK4 A0A182FEC0 A0A084W9M0 A0A182PTL8 A0A182HXV4 A0A182WS57 Q7PPU4 A0A182GLH9 A0A1B0GIH6 E2A775 A0A182IL33 E0VMH3 A0A182YBX8 A0A182NV16 A0A182K6S2 A0A1A9W2D3 A0A182QC09 A0A0C9R9E7 A0A182RYZ1 A0A0L0BRH5 A0A182WMI1 A0A0Q9WB25 A0A1A9Z260 B0WHK7 A0A1A9V040 A0A1A9Y7I5 A0A1B0GFF7 A0A1B0BG75 A0A0C9R2H4 Q7JVS8 B4P7L2 B4HN36 B4ME18 A0A1W4UI32 A0A182RY01 B4QB29 B4KQ21 A0A034WSM1 A0A0K8U7M5 F8J1K8 A0A088A650 B4J7X1 A0A2A3EJ03 B3NSN7 A0A182LHV6 B4H6C2 Q28WS2 B3MEQ2 A0A1I8PDP1 A0A0K8WGQ5 A0A1I8MJD1 B4MK79 A0A1S4FJS5 Q16YF4 A0A1B6I7F6 A0A0R3NPY6 U3U8X9 D6WTY0 A0A0J7KTA3 K7J443 A0A336LPL8 E2BHK2 A0A0M5IZK7 A0A1Y1MWW1 A0A139WAQ6 D6X512 A0A3B0J3V8 A0A1B6ECD2 T1J538
A0A212EX53 A0A2H1VIC9 A0A0N0P9E7 A0A0S1YD84 A0A2W1BU96 A0A194QJU0 B3XXL5 A0A2W1BS81 A0A194QVY6 A0A0S1YD68 A0A212FLW4 B3XXM1 A0A194QK98 A0A0S1YD70 A0A3S2LDK4 A0A182FEC0 A0A084W9M0 A0A182PTL8 A0A182HXV4 A0A182WS57 Q7PPU4 A0A182GLH9 A0A1B0GIH6 E2A775 A0A182IL33 E0VMH3 A0A182YBX8 A0A182NV16 A0A182K6S2 A0A1A9W2D3 A0A182QC09 A0A0C9R9E7 A0A182RYZ1 A0A0L0BRH5 A0A182WMI1 A0A0Q9WB25 A0A1A9Z260 B0WHK7 A0A1A9V040 A0A1A9Y7I5 A0A1B0GFF7 A0A1B0BG75 A0A0C9R2H4 Q7JVS8 B4P7L2 B4HN36 B4ME18 A0A1W4UI32 A0A182RY01 B4QB29 B4KQ21 A0A034WSM1 A0A0K8U7M5 F8J1K8 A0A088A650 B4J7X1 A0A2A3EJ03 B3NSN7 A0A182LHV6 B4H6C2 Q28WS2 B3MEQ2 A0A1I8PDP1 A0A0K8WGQ5 A0A1I8MJD1 B4MK79 A0A1S4FJS5 Q16YF4 A0A1B6I7F6 A0A0R3NPY6 U3U8X9 D6WTY0 A0A0J7KTA3 K7J443 A0A336LPL8 E2BHK2 A0A0M5IZK7 A0A1Y1MWW1 A0A139WAQ6 D6X512 A0A3B0J3V8 A0A1B6ECD2 T1J538
PDB
5N2S
E-value=0.00343092,
Score=93
Ontologies
GO
Topology
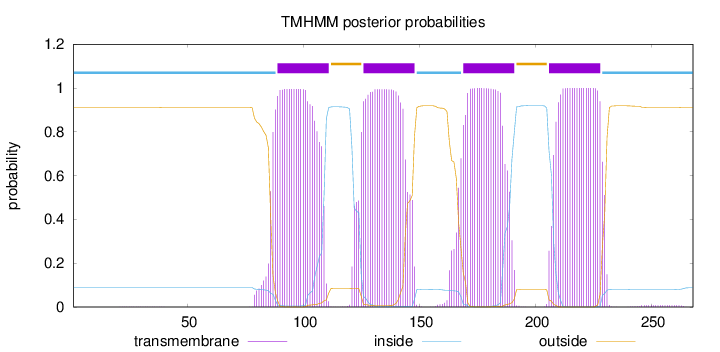
Length:
268
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
89.35966
Exp number, first 60 AAs:
0.00395
Total prob of N-in:
0.08923
inside
1 - 88
TMhelix
89 - 111
outside
112 - 125
TMhelix
126 - 148
inside
149 - 168
TMhelix
169 - 191
outside
192 - 205
TMhelix
206 - 228
inside
229 - 268
Population Genetic Test Statistics
Pi
3.998005
Theta
167.022715
Tajima's D
-0.853133
CLR
327.945379
CSRT
0.165591720413979
Interpretation
Uncertain
