Gene
KWMTBOMO11561
Pre Gene Modal
BGIBMGA004082
Annotation
PREDICTED:_tyrosine-protein_kinase_hopscotch_[Papilio_xuthus]
Full name
Tyrosine-protein kinase
+ More
Non-specific protein-tyrosine kinase
Non-specific protein-tyrosine kinase
Location in the cell
Nuclear Reliability : 2.473
Sequence
CDS
ATGGCAACAGTGGAAGATATAGTTAAAGTGGCCGTGGTGACCAATCGTACACCAAATAATGTAAGGTGTACTAGCACACTTACTGCAGAAGAATTGTGTATAATATTGTGTAAGAAATACAACATACCTCCTCTAACTCGTACATTATTTGCATTACGAGTTAAGGGTACTGATTATTTCCTCAAGGATAACTGTAAGGTCCTCTCGGGCTCTAGGGACTATGAACTGAGGATTCGTTTCAAGGTGCCAAGGTTAAGTCTTCTTATAACATTGGATGAGACCACATATGATTATTATTTCCAACAAGCAAGGAATGACGTCAATGAAAATAAAATACCTGAGATTAAGTATCCTGAACATAAGCAAGAATTACTCGGACTGGGGATCACAGATATGACCCGTGCAATAGCTGAAGAGAGATTACCACTGAATGATGTTATTCGAAATTACAAAAAATATATTCCTAAAGAGATTGCTAAAAGACATGGACCATTTCCTAAAAAGTATGCCATTGATTATCTACCGCAATTGTCTTCGGTCGGTCATAATGTGAATTACGTGAAGAATTTGTACTTACAACAGTTATACGCGTTAGCACCAAATTACCTCGCTGAGGAGTACGATAATGTTCTGTGGCAAAACGGCAGTGAGATGACGCCAGTGCGGGTGATGGTGGCACCTTTTCATCCGCACCATCCTGGAATACGACTTTACAACACTCTGAAACGTGAATGGTTCCATGTGTGCTCCGTCGAGGAGTTGATATATCTGACAAGGAACGGTGATAGTTCAGTGGAAGTGTCAAGAAGAGGCACTCCTGTGTTTCTCAAGTTCAAGTCCGAGGATCAGCTTTCGTCTTTTATTTCTGTCTGTGATGGATATTATAGGTTGACCGTAAAATGGACATTCAATCTCTCGAAAGACGATGAGACGCCTTCACTTAAAGAATTGCAAAGGATCAAATGTCATGGACCTGTTGGCGGTGCTTTCTCGTATCGGAAGTTGGAGGAGAAACGCGGAAAGAAACACGGGTGTTATATACTGAGGCAATGCCAGGACGAATATAACGTGTATTATTTGGACGTTTGTACGAAAACAAGCACAACAGAAACGTACAGAATAGAATTCAAAGGTCACTGCTACATATTCAGCACTGAAGAGTACTTTAGCATTGAACGGTTGGTCAGTTGCCATCAAAATCCCGAAGGAAGAATATACCTCAACGAATGCATACCTCCCTCTGAGTATGACAAATCTCAGTTGCTTTTGTGCGGGGAACCGGTGAAGAGGGGTGTCCGCATCGACCAGGCAGAATTACAGGAGATTCTGAGGGACAACAAGAGTCCCAGGTGCTTGCCTAACAAAGACATTTTATTGTATACAGGCTCCGAGAAAACGGGCTCAGAGAATATAACGGCGACATACAAAGCTCTGTGGAGGCTAGATGAGACAAAGAAATTGTTAGTTGCGTTCAAGACACTGCAACGGGAGAGGGCCAATGACTATTTAAAGGACTTTGTAGATCTCGCAAGCAAATGGGCGTGCATACAGTCTAGTTCTATAGTTAGGCTGTATGGAGTGACTTTGAGTTCCCCGACAGCAATGGTATTGGAATATCTACCGTACGGACCGTTTGATGATTATCTGAGAGAAAACGGCGACCGTATAACGCCACTGCACTTAAAGAAGGTAGCTGCTGGTCTGGCGAGGGCGCTGTGGGACCTGTCGGAGGCGGGCGTGGTGCACGGCTACATCAGGTGCCGGAAGCTGCTGCTGGCCGTGCTCGACGCGGACCGTATTCAAGTCAAGCTATCCGGCCCCACTCTGAGACAATACACGCAGCAGGATGTACATTGGATGCCTGTAGAATTTTTCGCAGACATGAACTTAGCGAAGAGATCGGTCATTGGAGACATTTGGGCTTTCGCTACAACGCTCTGGGAAGTATTCTCGTACGGACAATTGCCAACTGAAACCAACCCCGTTTTGACAGTTAGGAGCTACGAGATGGGCGACCGCCTGCTGCGGCCGGTCCGGTGCCCGGCGGAGGTGTGGGCGCTGGTGCGGCAGTGCTGGCACGCCGACCCGCTCCGTCCGCAGGAGATCATGCGGGACATGAACCACATGCTGTATAGAGAATATGTTCCTGTACATCAATATGAAGAACCTAAGATATCTCTTGATCACATAGAGCACGCAGAAACTGTGTCGAGTGATCGTTTTATTCCGAGCGAACTGAGTGACGCGGGAAGCAACAAATCTCTTATATCGGACAACAGCGTTCCCTCTATGAATGGCACTATGTCTGATCAGTATGACAATCCCTTCGCAGACAACAAAAGTTCGAACTCGCTGGACAGCATGGCGGCCCTGATGTACGGGCTCCCCAACGAGGTGTCGTCGGCGCGTAGCACGAGCAGCGCGGGCGGCGCCGCCACGCTGGGGCTCGACGACGCGCTGCACGACTGCGACGCCTGGGCCTCCCCCAGGCTCATGGAGTCCATCGTCTCGCAGGGGAAGACCTACCTCGTCACCTTGACCAAGAAGATCGGAAGCGGAAATTACGGTCACGTATTCAAAGGTTGGATGGAGCGTGACAATCAAGAATCTCACAGAAAAGAAGTGGCGATTAAGAAATTAACCCGACAAGCCTCCGAAAGGAACGGGACTCTGTATGAAGACTTCAAAAACGAATTGGAGATTATGAAGTCATTACAACACATTAACATAGTAGAGATACTGGGCTATGCTTGGGACCAGGGACCGGAAGTTCTAATAGTCATGGAGTATCTGGAAGAAGGTTCGCTTAACTACTACCTCAAATTCCAAGGCGAAAAGCTTAGGATATCGCATCTGCTCAAATATTGTAAGGACATAGCAACGGGTATGGACCACGTGTCTGCTAAAAACGTCGTGCATCGGGATTTGGCGACGCGGAATATTCTAGTGGTGAACAAATATCACGTTAAGATATCCGACTTCGGTTTGGCCAGAATCATTCCTAAAGAAGAAAACACGTACAGACTTAAGACTGAAAGACTGTTACCAATTAACTGGTACGCCCCCGAGTCGGCCGTCGAGCCGTGGCACTTCTCGACCAAAAGCGACGTGTGGTCGTACGGTGTAACCGCCTGGGAAATATTTACACGTGCCAGACAAGAAGTTCCTAAGTTCGACGTAGATCGGCCGAGGGAAAGAGCGTCGTGTTTTCAAATACCCGAAGAATGTCCCTCCGAGATATTCAGGTACCTCATGAAGGAGTGTTGGGCTCTCGATCCGAATTTACGGCCAAGGTTTATTGACCTAATACACACATGCAAGAGATTCATAGCGGAATATCAGTAA
Protein
MATVEDIVKVAVVTNRTPNNVRCTSTLTAEELCIILCKKYNIPPLTRTLFALRVKGTDYFLKDNCKVLSGSRDYELRIRFKVPRLSLLITLDETTYDYYFQQARNDVNENKIPEIKYPEHKQELLGLGITDMTRAIAEERLPLNDVIRNYKKYIPKEIAKRHGPFPKKYAIDYLPQLSSVGHNVNYVKNLYLQQLYALAPNYLAEEYDNVLWQNGSEMTPVRVMVAPFHPHHPGIRLYNTLKREWFHVCSVEELIYLTRNGDSSVEVSRRGTPVFLKFKSEDQLSSFISVCDGYYRLTVKWTFNLSKDDETPSLKELQRIKCHGPVGGAFSYRKLEEKRGKKHGCYILRQCQDEYNVYYLDVCTKTSTTETYRIEFKGHCYIFSTEEYFSIERLVSCHQNPEGRIYLNECIPPSEYDKSQLLLCGEPVKRGVRIDQAELQEILRDNKSPRCLPNKDILLYTGSEKTGSENITATYKALWRLDETKKLLVAFKTLQRERANDYLKDFVDLASKWACIQSSSIVRLYGVTLSSPTAMVLEYLPYGPFDDYLRENGDRITPLHLKKVAAGLARALWDLSEAGVVHGYIRCRKLLLAVLDADRIQVKLSGPTLRQYTQQDVHWMPVEFFADMNLAKRSVIGDIWAFATTLWEVFSYGQLPTETNPVLTVRSYEMGDRLLRPVRCPAEVWALVRQCWHADPLRPQEIMRDMNHMLYREYVPVHQYEEPKISLDHIEHAETVSSDRFIPSELSDAGSNKSLISDNSVPSMNGTMSDQYDNPFADNKSSNSLDSMAALMYGLPNEVSSARSTSSAGGAATLGLDDALHDCDAWASPRLMESIVSQGKTYLVTLTKKIGSGNYGHVFKGWMERDNQESHRKEVAIKKLTRQASERNGTLYEDFKNELEIMKSLQHINIVEILGYAWDQGPEVLIVMEYLEEGSLNYYLKFQGEKLRISHLLKYCKDIATGMDHVSAKNVVHRDLATRNILVVNKYHVKISDFGLARIIPKEENTYRLKTERLLPINWYAPESAVEPWHFSTKSDVWSYGVTAWEIFTRARQEVPKFDVDRPRERASCFQIPEECPSEIFRYLMKECWALDPNLRPRFIDLIHTCKRFIAEYQ
Summary
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
A0A194QQJ1
A0A212EMJ4
H9J3J5
A0A2W1BXH3
A0A194QVS9
A0A2H1V4N2
+ More
A0A1Y1K6R2 A0A1W4XLI2 E0VH77 A0A0C9QPB0 A0A1B6CRB6 A0A3R5SQ52 A0A0L7R787 A0A023EZE8 A0A088A072 A0A0A9ZBW4 K7J5I4 A0A2A3E5R4 A0A1B6DNP3 A0A026WQI1 A0A067QQD4 A0A2R7VYE5 A0A232EVJ5 A0A151IJH6 A0A151I1R5 A0A151JM27 F4X5P4 A0A151K168 A0A0J7KVQ2 A0A1D2NKP5 A0A226F4F6 A0A2S2Q101 A0A0L7L0P6 A0A1B6K188 X1WTY1 A0A2P8ZHB7 A0A1J1I4Z3 T1IKG8 A0A2R5LF78 E9FY31 A0A182HEY8 Q16LR4 A0A0P5UGE1 A0A0P5XHX5 A0A0P5IKW7 Q5QJV8 A0A0P5NZ21 A0A0P6FR99 E9J5E7 A0A0P5IHK5 A0A336MM17 A0A182QBQ8 A0A0P4ZTN8 A0A0P5ECJ7 A0A1L8DKE2 A0A182MMT3 A0A182NTX4 A0A0P5P6T5 A0A0H3WKI9 A0A2M4CSA0 A0A2M4A9H8 A0A0A6Z6I8 A0A2M4BAQ4 A0A1B0CSM3 A0A1A9XSL2 A0A1B0AQF5 A0A1A9UEI4 A0A240SX87 A0A182F4S9 A0A0P5C566 A0A0P6IGV6 A0A084W5U8 A0A0N8E9U0 A0A0P6EP44 A0A0P6EX11 A0A0P6B986 C6ZH45 A0A0N8EAE6 A0A2M4CTN1 A0A0N8CX53 A0A0P5D1I9 A0A0P4YVW0 A0A0P5VAA4 A0A1V9X6C9 A0A0K2UML1 T1JVU0 A0A2M4BCB6
A0A1Y1K6R2 A0A1W4XLI2 E0VH77 A0A0C9QPB0 A0A1B6CRB6 A0A3R5SQ52 A0A0L7R787 A0A023EZE8 A0A088A072 A0A0A9ZBW4 K7J5I4 A0A2A3E5R4 A0A1B6DNP3 A0A026WQI1 A0A067QQD4 A0A2R7VYE5 A0A232EVJ5 A0A151IJH6 A0A151I1R5 A0A151JM27 F4X5P4 A0A151K168 A0A0J7KVQ2 A0A1D2NKP5 A0A226F4F6 A0A2S2Q101 A0A0L7L0P6 A0A1B6K188 X1WTY1 A0A2P8ZHB7 A0A1J1I4Z3 T1IKG8 A0A2R5LF78 E9FY31 A0A182HEY8 Q16LR4 A0A0P5UGE1 A0A0P5XHX5 A0A0P5IKW7 Q5QJV8 A0A0P5NZ21 A0A0P6FR99 E9J5E7 A0A0P5IHK5 A0A336MM17 A0A182QBQ8 A0A0P4ZTN8 A0A0P5ECJ7 A0A1L8DKE2 A0A182MMT3 A0A182NTX4 A0A0P5P6T5 A0A0H3WKI9 A0A2M4CSA0 A0A2M4A9H8 A0A0A6Z6I8 A0A2M4BAQ4 A0A1B0CSM3 A0A1A9XSL2 A0A1B0AQF5 A0A1A9UEI4 A0A240SX87 A0A182F4S9 A0A0P5C566 A0A0P6IGV6 A0A084W5U8 A0A0N8E9U0 A0A0P6EP44 A0A0P6EX11 A0A0P6B986 C6ZH45 A0A0N8EAE6 A0A2M4CTN1 A0A0N8CX53 A0A0P5D1I9 A0A0P4YVW0 A0A0P5VAA4 A0A1V9X6C9 A0A0K2UML1 T1JVU0 A0A2M4BCB6
EC Number
2.7.10.2
Pubmed
EMBL
KQ458575
KPJ05796.1
AGBW02013823
OWR42696.1
BABH01021456
BABH01021457
+ More
BABH01021458 BABH01021459 KZ149917 PZC77917.1 KQ461073 KPJ09638.1 ODYU01000642 SOQ35759.1 GEZM01096910 JAV54487.1 DS235161 EEB12733.1 GBYB01002527 JAG72294.1 GEDC01021555 JAS15743.1 MG708321 QAB02861.1 KQ414642 KOC66704.1 GBBI01004104 JAC14608.1 GBHO01004309 GDHC01013917 JAG39295.1 JAQ04712.1 KZ288379 PBC26622.1 GEDC01010004 JAS27294.1 KK107128 QOIP01000006 EZA58280.1 RLU22133.1 KK853579 KDR06554.1 KK854124 PTY11710.1 NNAY01001988 OXU22374.1 KQ977412 KYN02881.1 KQ976553 KYM80826.1 KQ978966 KYN26896.1 GL888730 EGI58249.1 KQ981225 KYN44381.1 LBMM01002779 KMQ94378.1 LJIJ01000015 ODN05848.1 LNIX01000001 OXA64061.1 GGMS01001619 MBY70822.1 JTDY01003744 KOB69057.1 GECU01002773 JAT04934.1 ABLF02023796 ABLF02023800 PYGN01000057 PSN55899.1 CVRI01000038 CRK93974.1 JH430530 GGLE01004055 MBY08181.1 GL732527 EFX87846.1 JXUM01006685 JXUM01006686 JXUM01006687 JXUM01006688 JXUM01006689 JXUM01006690 JXUM01006691 KQ560220 KXJ83756.1 CH477896 EAT35265.2 GDIP01130422 JAL73292.1 GDIQ01209728 GDIP01071829 JAK41997.1 JAM31886.1 GDIQ01212580 JAK39145.1 AY278117 AAQ18517.1 GDIQ01135864 JAL15862.1 GDIQ01044105 JAN50632.1 GL768153 EFZ11955.1 GDIQ01214679 JAK37046.1 UFQT01001450 SSX30611.1 AXCN02000807 GDIP01208062 JAJ15340.1 GDIP01149980 JAJ73422.1 GFDF01007155 JAV06929.1 AXCM01003839 GDIQ01132818 JAL18908.1 KP310054 AKM12664.1 GGFL01004005 MBW68183.1 GGFK01004090 MBW37411.1 JQ340025 AFP74109.1 GGFJ01000975 MBW50116.1 AJWK01026286 AJWK01026287 AJWK01026288 JXJN01001847 GDIP01175820 JAJ47582.1 GDIQ01004670 JAN90067.1 ATLV01020694 KE525305 KFB45592.1 GDIQ01047748 JAN46989.1 GDIQ01059837 JAN34900.1 GDIQ01057345 JAN37392.1 GDIP01018460 JAM85255.1 EU877978 ACJ63722.1 GDIQ01046100 JAN48637.1 GGFL01004010 MBW68188.1 GDIP01090223 JAM13492.1 GDIP01162421 JAJ60981.1 GDIP01222909 JAJ00493.1 GDIP01104933 JAL98781.1 MNPL01022345 OQR69047.1 HACA01022148 CDW39509.1 CAEY01000797 GGFJ01001522 MBW50663.1
BABH01021458 BABH01021459 KZ149917 PZC77917.1 KQ461073 KPJ09638.1 ODYU01000642 SOQ35759.1 GEZM01096910 JAV54487.1 DS235161 EEB12733.1 GBYB01002527 JAG72294.1 GEDC01021555 JAS15743.1 MG708321 QAB02861.1 KQ414642 KOC66704.1 GBBI01004104 JAC14608.1 GBHO01004309 GDHC01013917 JAG39295.1 JAQ04712.1 KZ288379 PBC26622.1 GEDC01010004 JAS27294.1 KK107128 QOIP01000006 EZA58280.1 RLU22133.1 KK853579 KDR06554.1 KK854124 PTY11710.1 NNAY01001988 OXU22374.1 KQ977412 KYN02881.1 KQ976553 KYM80826.1 KQ978966 KYN26896.1 GL888730 EGI58249.1 KQ981225 KYN44381.1 LBMM01002779 KMQ94378.1 LJIJ01000015 ODN05848.1 LNIX01000001 OXA64061.1 GGMS01001619 MBY70822.1 JTDY01003744 KOB69057.1 GECU01002773 JAT04934.1 ABLF02023796 ABLF02023800 PYGN01000057 PSN55899.1 CVRI01000038 CRK93974.1 JH430530 GGLE01004055 MBY08181.1 GL732527 EFX87846.1 JXUM01006685 JXUM01006686 JXUM01006687 JXUM01006688 JXUM01006689 JXUM01006690 JXUM01006691 KQ560220 KXJ83756.1 CH477896 EAT35265.2 GDIP01130422 JAL73292.1 GDIQ01209728 GDIP01071829 JAK41997.1 JAM31886.1 GDIQ01212580 JAK39145.1 AY278117 AAQ18517.1 GDIQ01135864 JAL15862.1 GDIQ01044105 JAN50632.1 GL768153 EFZ11955.1 GDIQ01214679 JAK37046.1 UFQT01001450 SSX30611.1 AXCN02000807 GDIP01208062 JAJ15340.1 GDIP01149980 JAJ73422.1 GFDF01007155 JAV06929.1 AXCM01003839 GDIQ01132818 JAL18908.1 KP310054 AKM12664.1 GGFL01004005 MBW68183.1 GGFK01004090 MBW37411.1 JQ340025 AFP74109.1 GGFJ01000975 MBW50116.1 AJWK01026286 AJWK01026287 AJWK01026288 JXJN01001847 GDIP01175820 JAJ47582.1 GDIQ01004670 JAN90067.1 ATLV01020694 KE525305 KFB45592.1 GDIQ01047748 JAN46989.1 GDIQ01059837 JAN34900.1 GDIQ01057345 JAN37392.1 GDIP01018460 JAM85255.1 EU877978 ACJ63722.1 GDIQ01046100 JAN48637.1 GGFL01004010 MBW68188.1 GDIP01090223 JAM13492.1 GDIP01162421 JAJ60981.1 GDIP01222909 JAJ00493.1 GDIP01104933 JAL98781.1 MNPL01022345 OQR69047.1 HACA01022148 CDW39509.1 CAEY01000797 GGFJ01001522 MBW50663.1
Proteomes
UP000053268
UP000007151
UP000005204
UP000053240
UP000192223
UP000009046
+ More
UP000053825 UP000005203 UP000002358 UP000242457 UP000053097 UP000279307 UP000027135 UP000215335 UP000078542 UP000078540 UP000078492 UP000007755 UP000078541 UP000036403 UP000094527 UP000198287 UP000037510 UP000007819 UP000245037 UP000183832 UP000000305 UP000069940 UP000249989 UP000008820 UP000075886 UP000075883 UP000075884 UP000092461 UP000092443 UP000092460 UP000078200 UP000092445 UP000069272 UP000030765 UP000192247 UP000015104
UP000053825 UP000005203 UP000002358 UP000242457 UP000053097 UP000279307 UP000027135 UP000215335 UP000078542 UP000078540 UP000078492 UP000007755 UP000078541 UP000036403 UP000094527 UP000198287 UP000037510 UP000007819 UP000245037 UP000183832 UP000000305 UP000069940 UP000249989 UP000008820 UP000075886 UP000075883 UP000075884 UP000092461 UP000092443 UP000092460 UP000078200 UP000092445 UP000069272 UP000030765 UP000192247 UP000015104
PRIDE
Pfam
Interpro
IPR000719
Prot_kinase_dom
+ More
IPR020635 Tyr_kinase_cat_dom
IPR036860 SH2_dom_sf
IPR008266 Tyr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000299 FERM_domain
IPR019749 Band_41_domain
IPR019748 FERM_central
IPR016251 Tyr_kinase_non-rcpt_Jak/Tyk2
IPR041381 Jak1_PHL_dom
IPR041155 FERM_F1
IPR035963 FERM_2
IPR000980 SH2
IPR041046 FERM_F2
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR020635 Tyr_kinase_cat_dom
IPR036860 SH2_dom_sf
IPR008266 Tyr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000299 FERM_domain
IPR019749 Band_41_domain
IPR019748 FERM_central
IPR016251 Tyr_kinase_non-rcpt_Jak/Tyk2
IPR041381 Jak1_PHL_dom
IPR041155 FERM_F1
IPR035963 FERM_2
IPR000980 SH2
IPR041046 FERM_F2
IPR014352 FERM/acyl-CoA-bd_prot_sf
Gene 3D
ProteinModelPortal
A0A194QQJ1
A0A212EMJ4
H9J3J5
A0A2W1BXH3
A0A194QVS9
A0A2H1V4N2
+ More
A0A1Y1K6R2 A0A1W4XLI2 E0VH77 A0A0C9QPB0 A0A1B6CRB6 A0A3R5SQ52 A0A0L7R787 A0A023EZE8 A0A088A072 A0A0A9ZBW4 K7J5I4 A0A2A3E5R4 A0A1B6DNP3 A0A026WQI1 A0A067QQD4 A0A2R7VYE5 A0A232EVJ5 A0A151IJH6 A0A151I1R5 A0A151JM27 F4X5P4 A0A151K168 A0A0J7KVQ2 A0A1D2NKP5 A0A226F4F6 A0A2S2Q101 A0A0L7L0P6 A0A1B6K188 X1WTY1 A0A2P8ZHB7 A0A1J1I4Z3 T1IKG8 A0A2R5LF78 E9FY31 A0A182HEY8 Q16LR4 A0A0P5UGE1 A0A0P5XHX5 A0A0P5IKW7 Q5QJV8 A0A0P5NZ21 A0A0P6FR99 E9J5E7 A0A0P5IHK5 A0A336MM17 A0A182QBQ8 A0A0P4ZTN8 A0A0P5ECJ7 A0A1L8DKE2 A0A182MMT3 A0A182NTX4 A0A0P5P6T5 A0A0H3WKI9 A0A2M4CSA0 A0A2M4A9H8 A0A0A6Z6I8 A0A2M4BAQ4 A0A1B0CSM3 A0A1A9XSL2 A0A1B0AQF5 A0A1A9UEI4 A0A240SX87 A0A182F4S9 A0A0P5C566 A0A0P6IGV6 A0A084W5U8 A0A0N8E9U0 A0A0P6EP44 A0A0P6EX11 A0A0P6B986 C6ZH45 A0A0N8EAE6 A0A2M4CTN1 A0A0N8CX53 A0A0P5D1I9 A0A0P4YVW0 A0A0P5VAA4 A0A1V9X6C9 A0A0K2UML1 T1JVU0 A0A2M4BCB6
A0A1Y1K6R2 A0A1W4XLI2 E0VH77 A0A0C9QPB0 A0A1B6CRB6 A0A3R5SQ52 A0A0L7R787 A0A023EZE8 A0A088A072 A0A0A9ZBW4 K7J5I4 A0A2A3E5R4 A0A1B6DNP3 A0A026WQI1 A0A067QQD4 A0A2R7VYE5 A0A232EVJ5 A0A151IJH6 A0A151I1R5 A0A151JM27 F4X5P4 A0A151K168 A0A0J7KVQ2 A0A1D2NKP5 A0A226F4F6 A0A2S2Q101 A0A0L7L0P6 A0A1B6K188 X1WTY1 A0A2P8ZHB7 A0A1J1I4Z3 T1IKG8 A0A2R5LF78 E9FY31 A0A182HEY8 Q16LR4 A0A0P5UGE1 A0A0P5XHX5 A0A0P5IKW7 Q5QJV8 A0A0P5NZ21 A0A0P6FR99 E9J5E7 A0A0P5IHK5 A0A336MM17 A0A182QBQ8 A0A0P4ZTN8 A0A0P5ECJ7 A0A1L8DKE2 A0A182MMT3 A0A182NTX4 A0A0P5P6T5 A0A0H3WKI9 A0A2M4CSA0 A0A2M4A9H8 A0A0A6Z6I8 A0A2M4BAQ4 A0A1B0CSM3 A0A1A9XSL2 A0A1B0AQF5 A0A1A9UEI4 A0A240SX87 A0A182F4S9 A0A0P5C566 A0A0P6IGV6 A0A084W5U8 A0A0N8E9U0 A0A0P6EP44 A0A0P6EX11 A0A0P6B986 C6ZH45 A0A0N8EAE6 A0A2M4CTN1 A0A0N8CX53 A0A0P5D1I9 A0A0P4YVW0 A0A0P5VAA4 A0A1V9X6C9 A0A0K2UML1 T1JVU0 A0A2M4BCB6
PDB
4UEU
E-value=1.33623e-41,
Score=431
Ontologies
PATHWAY
GO
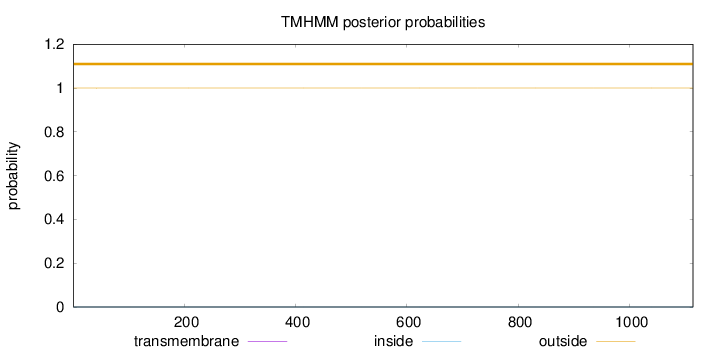
Topology
Length:
1114
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00359000000000001
Exp number, first 60 AAs:
0.00146
Total prob of N-in:
0.00017
outside
1 - 1114
Population Genetic Test Statistics
Pi
19.195598
Theta
169.657756
Tajima's D
0.458444
CLR
0.672545
CSRT
0.495175241237938
Interpretation
Uncertain
