Gene
KWMTBOMO11553
Annotation
PREDICTED:_uncharacterized_protein_LOC106132622_isoform_X1_[Amyelois_transitella]
Full name
Gustatory receptor
Location in the cell
PlasmaMembrane Reliability : 2.42
Sequence
CDS
ATGAGTACCAATAATGTGGATGTAATTAAGATGATCGGAAAATATTTAGATGTTTCAGTCGGGGCCGTTTGTTATAATACAGACAAAGTTCCCACTGCCGTACTTGAAAATAAAAATGTTGAGGGTTTTGTGACTATCATTTTGAGTATGGTTTCAAAATGTGGAACTGCAAGCAGTGAAGAGCAACGACTGCTAACCTACCAATGGTTAGAGTACATCTCCATGTTTAGTAATCAGGCTGTGGCAAATTCAACTTTTGCATTCAAATTCTTGCAAGAAATAAACAGAGCTCTACAAAGTAACACATATCTGACTGGCCAGTTTCTAACAATTGCTGATGTTGCATTATATTACATAGTGTATCCTTTACTGGAGCATATGTCTGTAGCAGAACGCGATGCCTTTGTTCACTTATGCAGATGGTCAAAACATATACAAGCACAACCGCGAGTTTGCCATAATCGTCCACCACTTCCATTGAATGCTGTGTCACTCTCGGTGCTGGCACCGGCAGTACACTGA
Protein
MSTNNVDVIKMIGKYLDVSVGAVCYNTDKVPTAVLENKNVEGFVTIILSMVSKCGTASSEEQRLLTYQWLEYISMFSNQAVANSTFAFKFLQEINRALQSNTYLTGQFLTIADVALYYIVYPLLEHMSVAERDAFVHLCRWSKHIQAQPRVCHNRPPLPLNAVSLSVLAPAVH
Summary
Description
Gustatory receptor which mediates acceptance or avoidance behavior, depending on its substrates.
Similarity
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
A0A194QJT0
A0A2W1BS62
A0A2A4JLY1
A0A2H1VN68
S4PB34
A0A3S2P7K1
+ More
A0A194QVZ6 A0A0L7LMN7 B4NMQ7 B4GHW3 Q28ZE1 A0A1I8MR84 A0A154P3V5 A0A1A9XXY3 A0A1A9VCA5 A0A034WDX4 A0A1A9X4H1 A0A1B0GAY1 A0A1B0AMC6 A0A1W4U9M8 A0A1B0ABN7 A0A1I8PQI9 A0A0K8UYY4 A0A0K8TSE2 B3NPN3 W8C5X4 B4P9N8 A0A0L0BPV6 A0A0M4EUS7 F4WE21 B4QIR2 B3MFG7 A0A195BBG9 B4MFQ3 B4I8P3 B4J9P4 Q8MKK1 A0A3B0JHZ0 Q8MTU4 A0A139WEF6 A0A2A3EJQ3 A0A0L7R1V4 A0A088A654 A0A0N1IU04 A0A1L8DS86 B4KRT8 A0A1W4WMQ6 A0A2P8XFB7 A0A1B0CK72 K7IQH1 A0A182WLZ1 A0A182PEN7 Q172H7 A0A084WCX1 A0A151IV25 E2A763 W5JSC2 A0A1S4FGA5 N6TL23 A0A2M4APY2 A0A2M3Z6W2 A0A3B3C975 A0A182HQI5 Q7Q7Z9 A0A182WYY2 A0A232F1D8 A0A2M4APV8 A0A2M3ZHT8 A0A151IN65 T1DHH2 A0A2M3ZHT5 A0A2M4C1G1 A0A182K4A6 A0A2M4AQH0 A0A026WQF2 A0A182N211 W5KF01 A0A1A8FMX9 A0A1L8DS91 A0A1A8GD01 A0A3B5L3I8 M3ZUW6 A0A0S7LTZ2 A0A182HET9 A0A3Q1IV25 B0XET7 A0A023EIE3 A0A3P9IKK6 A0A1L8E8J2 A0A1A8PWM3 A0A1A8QSL9 A0A2U9CYB4 A0A1A8LF80 A0A1A8Q336 A0A3Q2QA73 A0A087XSF5 A0A3B3WD12 F1QVE4
A0A194QVZ6 A0A0L7LMN7 B4NMQ7 B4GHW3 Q28ZE1 A0A1I8MR84 A0A154P3V5 A0A1A9XXY3 A0A1A9VCA5 A0A034WDX4 A0A1A9X4H1 A0A1B0GAY1 A0A1B0AMC6 A0A1W4U9M8 A0A1B0ABN7 A0A1I8PQI9 A0A0K8UYY4 A0A0K8TSE2 B3NPN3 W8C5X4 B4P9N8 A0A0L0BPV6 A0A0M4EUS7 F4WE21 B4QIR2 B3MFG7 A0A195BBG9 B4MFQ3 B4I8P3 B4J9P4 Q8MKK1 A0A3B0JHZ0 Q8MTU4 A0A139WEF6 A0A2A3EJQ3 A0A0L7R1V4 A0A088A654 A0A0N1IU04 A0A1L8DS86 B4KRT8 A0A1W4WMQ6 A0A2P8XFB7 A0A1B0CK72 K7IQH1 A0A182WLZ1 A0A182PEN7 Q172H7 A0A084WCX1 A0A151IV25 E2A763 W5JSC2 A0A1S4FGA5 N6TL23 A0A2M4APY2 A0A2M3Z6W2 A0A3B3C975 A0A182HQI5 Q7Q7Z9 A0A182WYY2 A0A232F1D8 A0A2M4APV8 A0A2M3ZHT8 A0A151IN65 T1DHH2 A0A2M3ZHT5 A0A2M4C1G1 A0A182K4A6 A0A2M4AQH0 A0A026WQF2 A0A182N211 W5KF01 A0A1A8FMX9 A0A1L8DS91 A0A1A8GD01 A0A3B5L3I8 M3ZUW6 A0A0S7LTZ2 A0A182HET9 A0A3Q1IV25 B0XET7 A0A023EIE3 A0A3P9IKK6 A0A1L8E8J2 A0A1A8PWM3 A0A1A8QSL9 A0A2U9CYB4 A0A1A8LF80 A0A1A8Q336 A0A3Q2QA73 A0A087XSF5 A0A3B3WD12 F1QVE4
Pubmed
26354079
28756777
23622113
26227816
17994087
15632085
+ More
25315136 25348373 26369729 24495485 17550304 26108605 21719571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 29403074 20075255 17510324 24438588 20798317 20920257 23761445 23537049 29451363 12364791 14747013 17210077 28648823 24508170 30249741 25329095 23542700 26483478 24945155 17554307 23594743
25315136 25348373 26369729 24495485 17550304 26108605 21719571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 29403074 20075255 17510324 24438588 20798317 20920257 23761445 23537049 29451363 12364791 14747013 17210077 28648823 24508170 30249741 25329095 23542700 26483478 24945155 17554307 23594743
EMBL
KQ458575
KPJ05787.1
KZ149917
PZC77909.1
NWSH01001023
PCG73067.1
+ More
ODYU01003225 SOQ41704.1 GAIX01006137 JAA86423.1 RSAL01000235 RVE43759.1 KQ461073 KPJ09647.1 JTDY01000574 KOB76619.1 CH964282 EDW85646.1 CH479183 EDW36083.1 CM000071 EAL25672.1 KQ434809 KZC06513.1 GAKP01006964 GAKP01006962 JAC51988.1 CCAG010004346 JXJN01000389 GDHF01020440 JAI31874.1 GDAI01000545 JAI17058.1 CH954179 EDV56824.1 GAMC01001452 JAC05104.1 CM000158 EDW92346.1 JRES01001567 KNC22003.1 CP012524 ALC41512.1 GL888102 EGI67459.1 CM000362 CM002911 EDX08390.1 KMY96053.1 CH902619 EDV36652.1 KQ976532 KYM81544.1 CH940667 EDW57224.1 CH480824 EDW56968.1 CH916367 EDW01458.1 AY113564 AE013599 AAM29569.1 AAM68253.1 OUUW01000001 SPP73009.1 AY071743 AAL49365.2 KQ971354 KYB26302.1 KZ288229 PBC31704.1 KQ414667 KOC64823.1 KQ435719 KOX78851.1 GFDF01004823 JAV09261.1 CH933808 EDW08358.1 PYGN01002348 PSN30697.1 AJWK01015852 AJWK01015853 CH477436 EAT40950.1 ATLV01022896 KE525337 KFB48065.1 KQ980926 KYN11396.1 GL437267 EFN70719.1 ADMH02000574 ETN65815.1 APGK01032677 KB740848 KB631977 ENN78653.1 ERL87579.1 GGFK01009509 MBW42830.1 GGFM01003515 MBW24266.1 APCN01004623 AAAB01008948 EAA10445.4 NNAY01001276 OXU24521.1 GGFK01009479 MBW42800.1 GGFM01007318 MBW28069.1 KQ976959 KYN06792.1 GAMD01002346 JAA99244.1 GGFM01007274 MBW28025.1 GGFJ01009850 MBW58991.1 GGFK01009703 MBW43024.1 KK107131 QOIP01000012 EZA58158.1 RLU16035.1 HAEB01013830 SBQ60357.1 GFDF01004837 JAV09247.1 HAEC01000829 SBQ68906.1 GBYX01150271 JAO92651.1 JXUM01036140 KQ561085 KXJ79756.1 DS232868 EDS26219.1 GAPW01004902 JAC08696.1 GFDG01003893 JAV14906.1 HAEI01003696 SBR85628.1 HAEH01012994 SBR96258.1 CP026263 AWP20829.1 HAEF01005796 SBR43178.1 HAEG01010643 SBR87667.1 AYCK01002860 CT573369 FO681333
ODYU01003225 SOQ41704.1 GAIX01006137 JAA86423.1 RSAL01000235 RVE43759.1 KQ461073 KPJ09647.1 JTDY01000574 KOB76619.1 CH964282 EDW85646.1 CH479183 EDW36083.1 CM000071 EAL25672.1 KQ434809 KZC06513.1 GAKP01006964 GAKP01006962 JAC51988.1 CCAG010004346 JXJN01000389 GDHF01020440 JAI31874.1 GDAI01000545 JAI17058.1 CH954179 EDV56824.1 GAMC01001452 JAC05104.1 CM000158 EDW92346.1 JRES01001567 KNC22003.1 CP012524 ALC41512.1 GL888102 EGI67459.1 CM000362 CM002911 EDX08390.1 KMY96053.1 CH902619 EDV36652.1 KQ976532 KYM81544.1 CH940667 EDW57224.1 CH480824 EDW56968.1 CH916367 EDW01458.1 AY113564 AE013599 AAM29569.1 AAM68253.1 OUUW01000001 SPP73009.1 AY071743 AAL49365.2 KQ971354 KYB26302.1 KZ288229 PBC31704.1 KQ414667 KOC64823.1 KQ435719 KOX78851.1 GFDF01004823 JAV09261.1 CH933808 EDW08358.1 PYGN01002348 PSN30697.1 AJWK01015852 AJWK01015853 CH477436 EAT40950.1 ATLV01022896 KE525337 KFB48065.1 KQ980926 KYN11396.1 GL437267 EFN70719.1 ADMH02000574 ETN65815.1 APGK01032677 KB740848 KB631977 ENN78653.1 ERL87579.1 GGFK01009509 MBW42830.1 GGFM01003515 MBW24266.1 APCN01004623 AAAB01008948 EAA10445.4 NNAY01001276 OXU24521.1 GGFK01009479 MBW42800.1 GGFM01007318 MBW28069.1 KQ976959 KYN06792.1 GAMD01002346 JAA99244.1 GGFM01007274 MBW28025.1 GGFJ01009850 MBW58991.1 GGFK01009703 MBW43024.1 KK107131 QOIP01000012 EZA58158.1 RLU16035.1 HAEB01013830 SBQ60357.1 GFDF01004837 JAV09247.1 HAEC01000829 SBQ68906.1 GBYX01150271 JAO92651.1 JXUM01036140 KQ561085 KXJ79756.1 DS232868 EDS26219.1 GAPW01004902 JAC08696.1 GFDG01003893 JAV14906.1 HAEI01003696 SBR85628.1 HAEH01012994 SBR96258.1 CP026263 AWP20829.1 HAEF01005796 SBR43178.1 HAEG01010643 SBR87667.1 AYCK01002860 CT573369 FO681333
Proteomes
UP000053268
UP000218220
UP000283053
UP000053240
UP000037510
UP000007798
+ More
UP000008744 UP000001819 UP000095301 UP000076502 UP000092443 UP000078200 UP000091820 UP000092444 UP000092460 UP000192221 UP000092445 UP000095300 UP000008711 UP000002282 UP000037069 UP000092553 UP000007755 UP000000304 UP000007801 UP000078540 UP000008792 UP000001292 UP000001070 UP000000803 UP000268350 UP000007266 UP000242457 UP000053825 UP000005203 UP000053105 UP000009192 UP000192223 UP000245037 UP000092461 UP000002358 UP000075920 UP000075885 UP000008820 UP000030765 UP000078492 UP000000311 UP000000673 UP000019118 UP000030742 UP000261560 UP000075840 UP000007062 UP000076407 UP000215335 UP000078542 UP000075881 UP000053097 UP000279307 UP000075884 UP000018467 UP000261380 UP000002852 UP000069940 UP000249989 UP000265040 UP000002320 UP000265200 UP000246464 UP000265000 UP000028760 UP000261480 UP000000437
UP000008744 UP000001819 UP000095301 UP000076502 UP000092443 UP000078200 UP000091820 UP000092444 UP000092460 UP000192221 UP000092445 UP000095300 UP000008711 UP000002282 UP000037069 UP000092553 UP000007755 UP000000304 UP000007801 UP000078540 UP000008792 UP000001292 UP000001070 UP000000803 UP000268350 UP000007266 UP000242457 UP000053825 UP000005203 UP000053105 UP000009192 UP000192223 UP000245037 UP000092461 UP000002358 UP000075920 UP000075885 UP000008820 UP000030765 UP000078492 UP000000311 UP000000673 UP000019118 UP000030742 UP000261560 UP000075840 UP000007062 UP000076407 UP000215335 UP000078542 UP000075881 UP000053097 UP000279307 UP000075884 UP000018467 UP000261380 UP000002852 UP000069940 UP000249989 UP000265040 UP000002320 UP000265200 UP000246464 UP000265000 UP000028760 UP000261480 UP000000437
Pfam
Interpro
IPR036282
Glutathione-S-Trfase_C_sf
+ More
IPR004046 GST_C
IPR010987 Glutathione-S-Trfase_C-like
IPR013604 7TM_chemorcpt
IPR020843 PKS_ER
IPR001214 SET_dom
IPR013154 ADH_N
IPR002893 Znf_MYND
IPR011990 TPR-like_helical_dom_sf
IPR011032 GroES-like_sf
IPR013149 ADH_C
IPR036291 NAD(P)-bd_dom_sf
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR036322 WD40_repeat_dom_sf
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR004046 GST_C
IPR010987 Glutathione-S-Trfase_C-like
IPR013604 7TM_chemorcpt
IPR020843 PKS_ER
IPR001214 SET_dom
IPR013154 ADH_N
IPR002893 Znf_MYND
IPR011990 TPR-like_helical_dom_sf
IPR011032 GroES-like_sf
IPR013149 ADH_C
IPR036291 NAD(P)-bd_dom_sf
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR036322 WD40_repeat_dom_sf
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A194QJT0
A0A2W1BS62
A0A2A4JLY1
A0A2H1VN68
S4PB34
A0A3S2P7K1
+ More
A0A194QVZ6 A0A0L7LMN7 B4NMQ7 B4GHW3 Q28ZE1 A0A1I8MR84 A0A154P3V5 A0A1A9XXY3 A0A1A9VCA5 A0A034WDX4 A0A1A9X4H1 A0A1B0GAY1 A0A1B0AMC6 A0A1W4U9M8 A0A1B0ABN7 A0A1I8PQI9 A0A0K8UYY4 A0A0K8TSE2 B3NPN3 W8C5X4 B4P9N8 A0A0L0BPV6 A0A0M4EUS7 F4WE21 B4QIR2 B3MFG7 A0A195BBG9 B4MFQ3 B4I8P3 B4J9P4 Q8MKK1 A0A3B0JHZ0 Q8MTU4 A0A139WEF6 A0A2A3EJQ3 A0A0L7R1V4 A0A088A654 A0A0N1IU04 A0A1L8DS86 B4KRT8 A0A1W4WMQ6 A0A2P8XFB7 A0A1B0CK72 K7IQH1 A0A182WLZ1 A0A182PEN7 Q172H7 A0A084WCX1 A0A151IV25 E2A763 W5JSC2 A0A1S4FGA5 N6TL23 A0A2M4APY2 A0A2M3Z6W2 A0A3B3C975 A0A182HQI5 Q7Q7Z9 A0A182WYY2 A0A232F1D8 A0A2M4APV8 A0A2M3ZHT8 A0A151IN65 T1DHH2 A0A2M3ZHT5 A0A2M4C1G1 A0A182K4A6 A0A2M4AQH0 A0A026WQF2 A0A182N211 W5KF01 A0A1A8FMX9 A0A1L8DS91 A0A1A8GD01 A0A3B5L3I8 M3ZUW6 A0A0S7LTZ2 A0A182HET9 A0A3Q1IV25 B0XET7 A0A023EIE3 A0A3P9IKK6 A0A1L8E8J2 A0A1A8PWM3 A0A1A8QSL9 A0A2U9CYB4 A0A1A8LF80 A0A1A8Q336 A0A3Q2QA73 A0A087XSF5 A0A3B3WD12 F1QVE4
A0A194QVZ6 A0A0L7LMN7 B4NMQ7 B4GHW3 Q28ZE1 A0A1I8MR84 A0A154P3V5 A0A1A9XXY3 A0A1A9VCA5 A0A034WDX4 A0A1A9X4H1 A0A1B0GAY1 A0A1B0AMC6 A0A1W4U9M8 A0A1B0ABN7 A0A1I8PQI9 A0A0K8UYY4 A0A0K8TSE2 B3NPN3 W8C5X4 B4P9N8 A0A0L0BPV6 A0A0M4EUS7 F4WE21 B4QIR2 B3MFG7 A0A195BBG9 B4MFQ3 B4I8P3 B4J9P4 Q8MKK1 A0A3B0JHZ0 Q8MTU4 A0A139WEF6 A0A2A3EJQ3 A0A0L7R1V4 A0A088A654 A0A0N1IU04 A0A1L8DS86 B4KRT8 A0A1W4WMQ6 A0A2P8XFB7 A0A1B0CK72 K7IQH1 A0A182WLZ1 A0A182PEN7 Q172H7 A0A084WCX1 A0A151IV25 E2A763 W5JSC2 A0A1S4FGA5 N6TL23 A0A2M4APY2 A0A2M3Z6W2 A0A3B3C975 A0A182HQI5 Q7Q7Z9 A0A182WYY2 A0A232F1D8 A0A2M4APV8 A0A2M3ZHT8 A0A151IN65 T1DHH2 A0A2M3ZHT5 A0A2M4C1G1 A0A182K4A6 A0A2M4AQH0 A0A026WQF2 A0A182N211 W5KF01 A0A1A8FMX9 A0A1L8DS91 A0A1A8GD01 A0A3B5L3I8 M3ZUW6 A0A0S7LTZ2 A0A182HET9 A0A3Q1IV25 B0XET7 A0A023EIE3 A0A3P9IKK6 A0A1L8E8J2 A0A1A8PWM3 A0A1A8QSL9 A0A2U9CYB4 A0A1A8LF80 A0A1A8Q336 A0A3Q2QA73 A0A087XSF5 A0A3B3WD12 F1QVE4
PDB
5Y6L
E-value=3.33563e-10,
Score=151
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
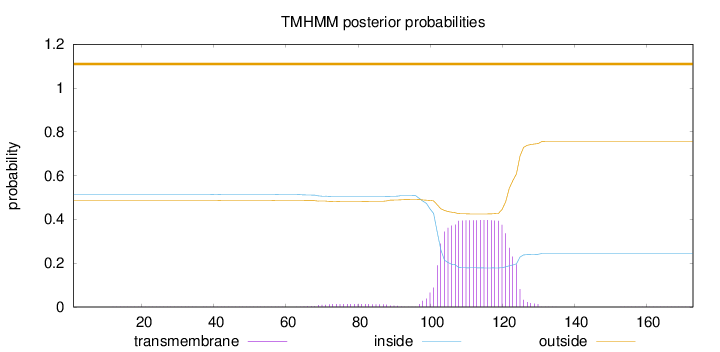
Length:
173
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.81373000000001
Exp number, first 60 AAs:
0.02157
Total prob of N-in:
0.51380
outside
1 - 173
Population Genetic Test Statistics
Pi
189.21799
Theta
174.038639
Tajima's D
0.274373
CLR
0.460837
CSRT
0.443577821108945
Interpretation
Uncertain
