Gene
KWMTBOMO11542
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Full name
Sodium/hydrogen exchanger
Location in the cell
Nuclear Reliability : 2.43
Sequence
CDS
ATGATTGCACACCGCAGAACCAGGCCCCTAGTCCCCGGCGGCCCCGCTATTCCTGGTTACGGTAGCGGCCATGGTAACGGCGGGGCAAGGGGTGCTAAGAATCCCCGGCAGAGATCCGGCTACCACCCCCCTTCACAACGACAACTGGCCCTGGTGACATACAACGTGCGCACTTTGAGGTTGGACGAGAAGCTAATAGAGCTGGAAGAAGAAATGAACAAGTTGCGCTGGGACGTTATTGGTTTATCGGAGATCCGAAGAGAGGGGGAGGATACGAAAACCTTACAGTCCGGCCACTTGTTCTACTACCGGGAGGGAGAACAGAAGTCCCAAGGTGGTGTTGGGTTTATCGTCCACAAATCTCTCATAAACAACATTGTGGCCATCGAAAGTGTGTCGAGTAGGGTAGTATACCTTGTCCTCAAAATCACAAAGCGGTATTCGCTGAAGATTGTACAGGTATACGCCCCATCGACGTCACATCCAGACGATGAAGTGGAAGCTACGTATGAGGACATTAGTAGGGCCATACGTAGCTCCCAATCACATTTTACCGTTGTTATGGGAGATTTCAATGCAAAACTGGGCCGTAGAGGCGATGATGAGTTGAAAGTGGGGCCATTTGGATTTGGGCAGCGGAATCCCAGAGGACAGATGCTGGCGGACTTTATGGAGAAGGAAGGACTCTTTATGATGAATTCTTTCTTCAAGAAGCCGCCACAACGGAAATGGACCTGGTTGAGTCCCGACGGTGTAACAAGAAATGAAATTGACTTCATCATGAGCACCAACAGACAGATATTCAACGATGTCTCTGTGATCAACAGTGTTAAAACAGGAAGTGATCACCGAATCGTCAGAGGCATGTTGAATATCAATGTCAGACTGGAAAGATCGCGTCTGATGAAGTCAACGCTTCGACCGTCGAATGCACAAATCCAAAACCCCGAGAGCTTTCAACTCGAATTGGCTAATCGCTTCGAATGGCTAAACAGCTGCGCATCAGTGGACGATGTAAACAGCAGGCTGGTAGAGACTGTCCGTACGGTAGGGTCGAAATTTTTCAAAACCCACCATACGAATAGACCTCAGAAACTATCCGACCGCACCCGAAACCTCATGGCTAGTCGACGTTTGATGACGCTGCAAACTTCGGAGGACGCTGAGTCATATAGGCAACTCAATAGACATATCATGAAGTCCTTGCGACACGACCTACGCTCCTTTAATACTCAGCGTATCAAAGAGACGATTGAGCGAAACCAAGGCTCTAAAGTGTTCGCAAGAGACGCTTCTATTGGGCAAAGCCAGCTGACGAAGCTGAAGACCGAAGATGGACGTGTAACGTCGAATAAGTCAGAGGTCCTACGGGAGATAGAGAGGTTCTACGGGCAGCTGTTTACCTCGGTCTCAGCTGAACCACAAGGTTTAGCTGCAGACCCTAGAGCCAAGCTAACCCGACACTACACCGAAGACATCCCGGACATCAGCCTGTACGAGATTAGGATGGCTCTCAAACAGCTTAAGAATAACAAGGCACCGGGCGAGGATGGAATTACTACGGAGCTTCTGAAAGCAGGCGGGACGCCGGTACTGAAAGTCCTTCAGAAGCTCTTCAATTCCTCTTTGCTCGATGGAAAACCGCCACAGGCATGGAACAGGGGTGTTGTGATCCTGTTCTTCAAAAAAGGTGACAAAACCTTATTGAAGAACTATAGGCCCATAGCACTGCTGAGCCATGTCTACAAGCTGTTCTCGAGAGTCATCACGAATCGTCTCGAACGCAGGTTTGATGACTTCCAGCCTACCGAACAAGCCGGTTTCCGAAAAGGCTACAGTACCATAGACCACATACATACGCTGCGGCAGATTGTACAGAAGACCGAAGAGTACAATCGGCCATTATGCTTAGCGTTTGTGGACTATGAGAAAGCCTTTGATTCCGTCGAAACATGGGCGGTCCTTAGATCTCTCCAGCGGTGCCGTATTGACCACCGGTACATCGAAGTATTGAAGTGTTTGTATAATAACGCCACCATGTCAGTCCGAGTACAGGAGCATTGCACGAAGGAAATCCCTGTAAAGAGAGGAGTGAGACAGGGAGATGTGATATCTCCGAAACTGTTTATCACTGCTCTGGAGGATTCCTTCAAGCTTCTGGAATGGCAAGGACTTGGCATCAATATTAACGGCGAATACATCACTCATCTTCGGTTTGCCGATGACATCGTGGTCATGGCAGAGTCGCTGGAAGATTTAGGGCGTATGCTCGGTGACCTCAGTAGGGTTTCTCAACAACTCTACTCTTGA
Protein
MIAHRRTRPLVPGGPAIPGYGSGHGNGGARGAKNPRQRSGYHPPSQRQLALVTYNVRTLRLDEKLIELEEEMNKLRWDVIGLSEIRREGEDTKTLQSGHLFYYREGEQKSQGGVGFIVHKSLINNIVAIESVSSRVVYLVLKITKRYSLKIVQVYAPSTSHPDDEVEATYEDISRAIRSSQSHFTVVMGDFNAKLGRRGDDELKVGPFGFGQRNPRGQMLADFMEKEGLFMMNSFFKKPPQRKWTWLSPDGVTRNEIDFIMSTNRQIFNDVSVINSVKTGSDHRIVRGMLNINVRLERSRLMKSTLRPSNAQIQNPESFQLELANRFEWLNSCASVDDVNSRLVETVRTVGSKFFKTHHTNRPQKLSDRTRNLMASRRLMTLQTSEDAESYRQLNRHIMKSLRHDLRSFNTQRIKETIERNQGSKVFARDASIGQSQLTKLKTEDGRVTSNKSEVLREIERFYGQLFTSVSAEPQGLAADPRAKLTRHYTEDIPDISLYEIRMALKQLKNNKAPGEDGITTELLKAGGTPVLKVLQKLFNSSLLDGKPPQAWNRGVVILFFKKGDKTLLKNYRPIALLSHVYKLFSRVITNRLERRFDDFQPTEQAGFRKGYSTIDHIHTLRQIVQKTEEYNRPLCLAFVDYEKAFDSVETWAVLRSLQRCRIDHRYIEVLKCLYNNATMSVRVQEHCTKEIPVKRGVRQGDVISPKLFITALEDSFKLLEWQGLGININGEYITHLRFADDIVVMAESLEDLGRMLGDLSRVSQQLYS
Summary
Similarity
Belongs to the monovalent cation:proton antiporter 1 (CPA1) transporter (TC 2.A.36) family.
Feature
chain Sodium/hydrogen exchanger
Uniprot
D7F158
D7F159
D7F164
D7F157
A0A3S2N805
D7F165
+ More
D7F160 D7F166 A0A2H1VS24 A0A2W1BZV1 A0A1S3HCU5 A0A0N5C0B1 A0A0K0FAW8 A0A023EWU8 A0A0P4VWD0 E3NV79 A0A016V4Q8 E3MH09 D7F177 A0A016TK11 A0A016SG46 K7HC62 K7HC63 D7F176 E3NC15 A0A016W4R1 K7IG35 K7IBZ6 K7IBZ5 D7F179 K7HC61 A0A2R2MNC1 A0A2H2J8I4 A0A016V9Q8 K7I2A3 A0A2H1WF27 A0A016V6A2 E3NJQ7 K7ICT2 K7ICT1 K7IG36 K7H0Q9 A0A016S1D2 K7H0R0 A0A016WCB8 A0A016WPY4 A0A2H2JFN0 K7HDM3 K7HDM2 A0A016ULU9 K7I2A2 K7I2A1 Q9TZX4 K7H0Q8 A0A016SPR9 A0A016V419 A0A016TIG5 A0A016WFS9 A0A016WUN2 A0A016U352 A0A016U3L3 A0A016SVA2 A0A016TQJ1 A0A016W1X2 A0A016U6B7 A0A016WYN9 A0A016UA47 A0A016SCL7 A0A016VLP0 A0A016T4C4 A0A016SW37 A0A016WRX9 A0A016UY78 A0A016X207 A0A016V007 A0A016SY13 A0A016U123 A0A016SCZ8 A0A016VT25 A0A023EXV2 A0A016UJS4 A0A016WW86 A0A016TX70 A0A016T9D3 A0A016SXL6 A0A016S7F6 A0A016WPX9 W6NF02 A0A016T9A5 A0A016WE57 A0A016SQY7
D7F160 D7F166 A0A2H1VS24 A0A2W1BZV1 A0A1S3HCU5 A0A0N5C0B1 A0A0K0FAW8 A0A023EWU8 A0A0P4VWD0 E3NV79 A0A016V4Q8 E3MH09 D7F177 A0A016TK11 A0A016SG46 K7HC62 K7HC63 D7F176 E3NC15 A0A016W4R1 K7IG35 K7IBZ6 K7IBZ5 D7F179 K7HC61 A0A2R2MNC1 A0A2H2J8I4 A0A016V9Q8 K7I2A3 A0A2H1WF27 A0A016V6A2 E3NJQ7 K7ICT2 K7ICT1 K7IG36 K7H0Q9 A0A016S1D2 K7H0R0 A0A016WCB8 A0A016WPY4 A0A2H2JFN0 K7HDM3 K7HDM2 A0A016ULU9 K7I2A2 K7I2A1 Q9TZX4 K7H0Q8 A0A016SPR9 A0A016V419 A0A016TIG5 A0A016WFS9 A0A016WUN2 A0A016U352 A0A016U3L3 A0A016SVA2 A0A016TQJ1 A0A016W1X2 A0A016U6B7 A0A016WYN9 A0A016UA47 A0A016SCL7 A0A016VLP0 A0A016T4C4 A0A016SW37 A0A016WRX9 A0A016UY78 A0A016X207 A0A016V007 A0A016SY13 A0A016U123 A0A016SCZ8 A0A016VT25 A0A023EXV2 A0A016UJS4 A0A016WW86 A0A016TX70 A0A016T9D3 A0A016SXL6 A0A016S7F6 A0A016WPX9 W6NF02 A0A016T9A5 A0A016WE57 A0A016SQY7
EMBL
FJ265543
ADI61811.1
FJ265544
ADI61812.1
FJ265549
ADI61817.1
+ More
FJ265542 ADI61810.1 RSAL01001818 RVE40675.1 FJ265550 ADI61818.1 FJ265545 ADI61813.1 FJ265551 ADI61819.1 ODYU01004088 SOQ43598.1 KZ149907 PZC78256.1 GBBI01004877 JAC13835.1 GDKW01001596 JAI54999.1 DS270843 EFO97547.1 JARK01001354 EYC21713.1 DS268444 EFP01729.1 FJ265562 ADI61830.1 JARK01001676 JARK01001446 JARK01001433 EYB83199.1 EYC01126.1 EYC02928.1 JARK01001565 EYB89683.1 FJ265561 ADI61829.1 DS268591 EFO92565.1 JARK01001337 EYC34625.1 FJ265564 ADI61832.1 JARK01001351 EYC23458.1 ODYU01008249 SOQ51673.1 JARK01001353 EYC22263.1 DS268752 EFP01106.1 JARK01001655 EYB84321.1 JARK01000388 EYC37469.1 JARK01000173 EYC41342.1 JARK01001370 EYC16150.1 AF054983 AAC72298.1 JARK01001531 EYB92324.1 EYC21986.1 JARK01001435 EYC02496.1 JARK01000302 EYC38674.1 JARK01000109 EYC42952.1 JARK01001395 EYC09729.1 EYC09730.1 JARK01001505 EYB94618.1 JARK01001421 EYC04912.1 JARK01001338 EYC33297.1 JARK01001389 EYC10874.1 JARK01000063 EYC44387.1 JARK01001384 EYC12189.1 JARK01001628 JARK01001583 JARK01001528 JARK01001506 JARK01001470 JARK01001467 JARK01001463 JARK01001457 JARK01001432 JARK01001420 JARK01001403 JARK01001371 JARK01001369 JARK01001360 JARK01001349 JARK01000603 JARK01000080 EYB85741.1 EYB88438.1 EYB92544.1 EYB94540.1 EYB98080.1 EYB98468.1 EYB98899.1 EYB99568.1 EYC03088.1 EYC05014.1 EYC08153.1 EYC15852.1 EYC16408.1 EYC19456.1 EYC22445.1 EYC24734.1 EYC24778.1 EYC24866.1 EYC33488.1 EYC35672.1 EYC43798.1 JARK01001344 EYC28216.1 JARK01001475 EYB97550.1 EYB94617.1 JARK01000132 EYC42415.1 JARK01001359 EYC20105.1 JARK01000008 EYC46079.1 JARK01001357 EYC20551.1 JARK01001497 EYB95322.1 JARK01001401 EYC08686.1 JARK01001581 EYB88505.1 JARK01001340 EYC30744.1 GBBI01004876 JAC13836.1 JARK01001372 EYC15614.1 JARK01000078 EYC43871.1 JARK01001407 EYC07212.1 JARK01001459 EYB99276.1 JARK01001500 EYB95049.1 JARK01001612 EYB86585.1 JARK01000161 EYC41641.1 CAVP010059514 CDL95876.1 EYB99275.1 JARK01000343 EYC38099.1 JARK01001524 EYB92970.1
FJ265542 ADI61810.1 RSAL01001818 RVE40675.1 FJ265550 ADI61818.1 FJ265545 ADI61813.1 FJ265551 ADI61819.1 ODYU01004088 SOQ43598.1 KZ149907 PZC78256.1 GBBI01004877 JAC13835.1 GDKW01001596 JAI54999.1 DS270843 EFO97547.1 JARK01001354 EYC21713.1 DS268444 EFP01729.1 FJ265562 ADI61830.1 JARK01001676 JARK01001446 JARK01001433 EYB83199.1 EYC01126.1 EYC02928.1 JARK01001565 EYB89683.1 FJ265561 ADI61829.1 DS268591 EFO92565.1 JARK01001337 EYC34625.1 FJ265564 ADI61832.1 JARK01001351 EYC23458.1 ODYU01008249 SOQ51673.1 JARK01001353 EYC22263.1 DS268752 EFP01106.1 JARK01001655 EYB84321.1 JARK01000388 EYC37469.1 JARK01000173 EYC41342.1 JARK01001370 EYC16150.1 AF054983 AAC72298.1 JARK01001531 EYB92324.1 EYC21986.1 JARK01001435 EYC02496.1 JARK01000302 EYC38674.1 JARK01000109 EYC42952.1 JARK01001395 EYC09729.1 EYC09730.1 JARK01001505 EYB94618.1 JARK01001421 EYC04912.1 JARK01001338 EYC33297.1 JARK01001389 EYC10874.1 JARK01000063 EYC44387.1 JARK01001384 EYC12189.1 JARK01001628 JARK01001583 JARK01001528 JARK01001506 JARK01001470 JARK01001467 JARK01001463 JARK01001457 JARK01001432 JARK01001420 JARK01001403 JARK01001371 JARK01001369 JARK01001360 JARK01001349 JARK01000603 JARK01000080 EYB85741.1 EYB88438.1 EYB92544.1 EYB94540.1 EYB98080.1 EYB98468.1 EYB98899.1 EYB99568.1 EYC03088.1 EYC05014.1 EYC08153.1 EYC15852.1 EYC16408.1 EYC19456.1 EYC22445.1 EYC24734.1 EYC24778.1 EYC24866.1 EYC33488.1 EYC35672.1 EYC43798.1 JARK01001344 EYC28216.1 JARK01001475 EYB97550.1 EYB94617.1 JARK01000132 EYC42415.1 JARK01001359 EYC20105.1 JARK01000008 EYC46079.1 JARK01001357 EYC20551.1 JARK01001497 EYB95322.1 JARK01001401 EYC08686.1 JARK01001581 EYB88505.1 JARK01001340 EYC30744.1 GBBI01004876 JAC13836.1 JARK01001372 EYC15614.1 JARK01000078 EYC43871.1 JARK01001407 EYC07212.1 JARK01001459 EYB99276.1 JARK01001500 EYB95049.1 JARK01001612 EYB86585.1 JARK01000161 EYC41641.1 CAVP010059514 CDL95876.1 EYB99275.1 JARK01000343 EYC38099.1 JARK01001524 EYB92970.1
Proteomes
Pfam
Interpro
IPR005135
Endo/exonuclease/phosphatase
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR016186 C-type_lectin-like/link_sf
IPR016187 CTDL_fold
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR002213 UDP_glucos_trans
IPR001507 ZP_dom
IPR007741 Ribosome/NADH_DH
IPR036249 Thioredoxin-like_sf
IPR019430 7TM_GPCR_serpentine_rcpt_Srx
IPR017452 GPCR_Rhodpsn_7TM
IPR018422 Cation/H_exchanger_CPA1
IPR006153 Cation/H_exchanger
IPR004709 NaH_exchanger
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR016186 C-type_lectin-like/link_sf
IPR016187 CTDL_fold
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR002213 UDP_glucos_trans
IPR001507 ZP_dom
IPR007741 Ribosome/NADH_DH
IPR036249 Thioredoxin-like_sf
IPR019430 7TM_GPCR_serpentine_rcpt_Srx
IPR017452 GPCR_Rhodpsn_7TM
IPR018422 Cation/H_exchanger_CPA1
IPR006153 Cation/H_exchanger
IPR004709 NaH_exchanger
Gene 3D
ProteinModelPortal
D7F158
D7F159
D7F164
D7F157
A0A3S2N805
D7F165
+ More
D7F160 D7F166 A0A2H1VS24 A0A2W1BZV1 A0A1S3HCU5 A0A0N5C0B1 A0A0K0FAW8 A0A023EWU8 A0A0P4VWD0 E3NV79 A0A016V4Q8 E3MH09 D7F177 A0A016TK11 A0A016SG46 K7HC62 K7HC63 D7F176 E3NC15 A0A016W4R1 K7IG35 K7IBZ6 K7IBZ5 D7F179 K7HC61 A0A2R2MNC1 A0A2H2J8I4 A0A016V9Q8 K7I2A3 A0A2H1WF27 A0A016V6A2 E3NJQ7 K7ICT2 K7ICT1 K7IG36 K7H0Q9 A0A016S1D2 K7H0R0 A0A016WCB8 A0A016WPY4 A0A2H2JFN0 K7HDM3 K7HDM2 A0A016ULU9 K7I2A2 K7I2A1 Q9TZX4 K7H0Q8 A0A016SPR9 A0A016V419 A0A016TIG5 A0A016WFS9 A0A016WUN2 A0A016U352 A0A016U3L3 A0A016SVA2 A0A016TQJ1 A0A016W1X2 A0A016U6B7 A0A016WYN9 A0A016UA47 A0A016SCL7 A0A016VLP0 A0A016T4C4 A0A016SW37 A0A016WRX9 A0A016UY78 A0A016X207 A0A016V007 A0A016SY13 A0A016U123 A0A016SCZ8 A0A016VT25 A0A023EXV2 A0A016UJS4 A0A016WW86 A0A016TX70 A0A016T9D3 A0A016SXL6 A0A016S7F6 A0A016WPX9 W6NF02 A0A016T9A5 A0A016WE57 A0A016SQY7
D7F160 D7F166 A0A2H1VS24 A0A2W1BZV1 A0A1S3HCU5 A0A0N5C0B1 A0A0K0FAW8 A0A023EWU8 A0A0P4VWD0 E3NV79 A0A016V4Q8 E3MH09 D7F177 A0A016TK11 A0A016SG46 K7HC62 K7HC63 D7F176 E3NC15 A0A016W4R1 K7IG35 K7IBZ6 K7IBZ5 D7F179 K7HC61 A0A2R2MNC1 A0A2H2J8I4 A0A016V9Q8 K7I2A3 A0A2H1WF27 A0A016V6A2 E3NJQ7 K7ICT2 K7ICT1 K7IG36 K7H0Q9 A0A016S1D2 K7H0R0 A0A016WCB8 A0A016WPY4 A0A2H2JFN0 K7HDM3 K7HDM2 A0A016ULU9 K7I2A2 K7I2A1 Q9TZX4 K7H0Q8 A0A016SPR9 A0A016V419 A0A016TIG5 A0A016WFS9 A0A016WUN2 A0A016U352 A0A016U3L3 A0A016SVA2 A0A016TQJ1 A0A016W1X2 A0A016U6B7 A0A016WYN9 A0A016UA47 A0A016SCL7 A0A016VLP0 A0A016T4C4 A0A016SW37 A0A016WRX9 A0A016UY78 A0A016X207 A0A016V007 A0A016SY13 A0A016U123 A0A016SCZ8 A0A016VT25 A0A023EXV2 A0A016UJS4 A0A016WW86 A0A016TX70 A0A016T9D3 A0A016SXL6 A0A016S7F6 A0A016WPX9 W6NF02 A0A016T9A5 A0A016WE57 A0A016SQY7
PDB
6AR3
E-value=4.57337e-05,
Score=115
Ontologies
PANTHER
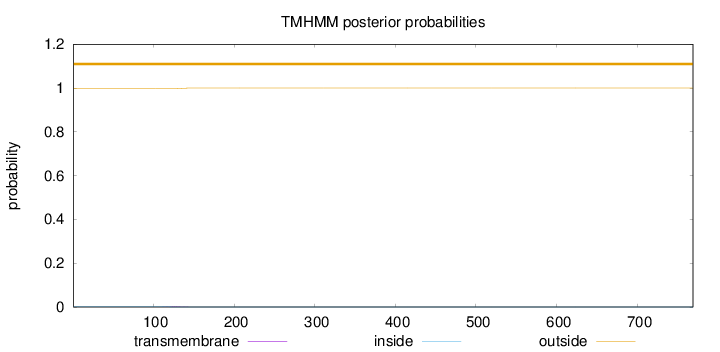
Topology
Length:
769
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0327300000000001
Exp number, first 60 AAs:
0.00223
Total prob of N-in:
0.00172
outside
1 - 769
Population Genetic Test Statistics
Pi
39.127735
Theta
30.384448
Tajima's D
-1.099704
CLR
282.244952
CSRT
0.117694115294235
Interpretation
Uncertain
