Gene
KWMTBOMO11541
Pre Gene Modal
BGIBMGA004040
Annotation
chemosensory_protein_8_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.069
Sequence
CDS
ATGAAGACGATCTTGATTTTATGCGCGTTAGTATCGGTCGTTGTTTGCCGACCGGAAGAATACTACAGCAGTCAATATGATAATTTTGACGTCGAGCAGCTGGTGGGCAACTTGCGCCTCTTGAAGAACTATGCCAAGTGCTTCCTCGACCAAGGCCCCTGTACCGCCGAAGGAACAGAATTTAAGAAAAGAATCCCCGAGGCGCTTAGAACTAAATGCGCCAAATGCAACCCGAAACAGCGTCATCTGATCCGTACAGTGGTAAAGGCTTTCCAGACTAAACTGCCTGACCTGTGGGAAGAGCTTGCCATAAAGGAGGACCCCAAGGGCCAATACAAACATGAATTTACTGCTTTCATCAACGCCATGGACTAA
Protein
MKTILILCALVSVVVCRPEEYYSSQYDNFDVEQLVGNLRLLKNYAKCFLDQGPCTAEGTEFKKRIPEALRTKCAKCNPKQRHLIRTVVKAFQTKLPDLWEELAIKEDPKGQYKHEFTAFINAMD
Summary
Uniprot
Q3LB97
A0A1B3P5K5
A0A1B4ZBJ1
A0A345BEN2
A0A3G6V6M0
E7EC45
+ More
A0A0C4KD22 A0A172QDN4 A0A2K8GKZ4 A0A076E960 A0A1L2BLB2 A0A1V1WBX1 A0A1W5LB26 A0A0K1DCX9 A0A0P0EW15 A0A1E1W9K9 M4XYS2 A0A290U466 S4PC20 U5KD25 A8QWQ6 A0A194QJL9 A0A1Z2R8Q9 A0A2W1BU58 A0A076E970 A0A0A0VG76 A0A212F0D7 A0A194QW11 A0A3Q9NAY6 A0A2A4JRD7 A0A386H8V2 A0A0L7LEQ2 A0A345BEN0 Q3LB44 A2TIK5 A0A194R1E1 A0A212F7W2 I4DIM2 A0A194QK61 A8QWQ3 A0A076E964 A0A212F0I9 A0A076E7E7 A0A2K8GL22 Q4VR77 A0A345BEN5 A0A0L7LL43 A0A0A1CR78 Q0MRJ2 A0A0C4KCE8 A0A2H1UZS0 Q3LB41 A0A3B0EM79 A0A1B4ZBL4 D2SNX2 A0A172QDP1 A0A1E1W7L4 H6TDX9 A0A2A4JQ79 A0A1B3P5L7 A0A1L2BL66 M4Y081 A0A1W5LAR7 U5PZS2 J9VHK5 A0A2H1V9F0 A0A0K8TVZ7 A0A3G6V6Z3 A0A0K8TW01 U5PZD9 Q3LB98 A0A0U2SQP9 A0A0M4FCQ3 A0A0U3AMZ7 U5Q2S2 A0A076E5V2 A0A1V1WC03 A0A386H9C5 A0A0C4K658 A0A0P0EM27 A0A109ZT31 A0A345BEP5 A0A076ED87 A0A3G6V6L7 A0A2A4JPI3 A0A1B4ZBL1 D2SNM8 A0A0K1DDI7 U5PZ61 A0A345BEN1 M4VTH9 A0A0L7RK18 A0A212FBG3 Q8I6X7 A0A345F0U4 A0A2W1BS20 A0A2K8GL19 A0A0K1DDT3 Q3LB48 A0A0U3T5N0
A0A0C4KD22 A0A172QDN4 A0A2K8GKZ4 A0A076E960 A0A1L2BLB2 A0A1V1WBX1 A0A1W5LB26 A0A0K1DCX9 A0A0P0EW15 A0A1E1W9K9 M4XYS2 A0A290U466 S4PC20 U5KD25 A8QWQ6 A0A194QJL9 A0A1Z2R8Q9 A0A2W1BU58 A0A076E970 A0A0A0VG76 A0A212F0D7 A0A194QW11 A0A3Q9NAY6 A0A2A4JRD7 A0A386H8V2 A0A0L7LEQ2 A0A345BEN0 Q3LB44 A2TIK5 A0A194R1E1 A0A212F7W2 I4DIM2 A0A194QK61 A8QWQ3 A0A076E964 A0A212F0I9 A0A076E7E7 A0A2K8GL22 Q4VR77 A0A345BEN5 A0A0L7LL43 A0A0A1CR78 Q0MRJ2 A0A0C4KCE8 A0A2H1UZS0 Q3LB41 A0A3B0EM79 A0A1B4ZBL4 D2SNX2 A0A172QDP1 A0A1E1W7L4 H6TDX9 A0A2A4JQ79 A0A1B3P5L7 A0A1L2BL66 M4Y081 A0A1W5LAR7 U5PZS2 J9VHK5 A0A2H1V9F0 A0A0K8TVZ7 A0A3G6V6Z3 A0A0K8TW01 U5PZD9 Q3LB98 A0A0U2SQP9 A0A0M4FCQ3 A0A0U3AMZ7 U5Q2S2 A0A076E5V2 A0A1V1WC03 A0A386H9C5 A0A0C4K658 A0A0P0EM27 A0A109ZT31 A0A345BEP5 A0A076ED87 A0A3G6V6L7 A0A2A4JPI3 A0A1B4ZBL1 D2SNM8 A0A0K1DDI7 U5PZ61 A0A345BEN1 M4VTH9 A0A0L7RK18 A0A212FBG3 Q8I6X7 A0A345F0U4 A0A2W1BS20 A0A2K8GL19 A0A0K1DDT3 Q3LB48 A0A0U3T5N0
Pubmed
EMBL
BABH01021434
DQ855514
AJ973409
ABH88201.1
CAJ01456.1
KX655941
+ More
AOG12890.1 LC027703 BAV56806.1 MG788188 AXF48706.1 MH027976 AZB49394.1 HQ649755 ADV36661.1 KF026051 AHX37220.1 KT357396 AND82444.1 KX585414 ARO70307.1 KF487616 AII01014.1 KT282984 ALS03841.1 GENK01000048 JAV45865.1 KT991357 ANA75023.1 KP729031 AKT26489.1 KT261677 ALJ30217.1 GDQN01007391 GDQN01007068 JAT83663.1 JAT83986.1 KC507180 AGI37363.1 KX954372 ATD12156.1 GAIX01008045 JAA84515.1 JX863697 AGR39572.1 AB260121 BAF91716.1 KQ458575 KPJ05768.1 KY815027 ASA40088.1 KZ149917 PZC77881.1 KF487636 AII01034.1 KM236063 AIW65100.1 AGBW02011101 OWR47215.1 KQ461073 KPJ09662.1 MH050646 AZT78915.1 NWSH01000827 PCG73980.1 MG546594 AYD42217.1 JTDY01001439 KOB73859.1 MG788186 AXF48704.1 AJ973462 CAJ01509.1 EF202829 FJ948817 ABM92664.1 ACR43877.1 KPJ09666.1 AGBW02009828 OWR49808.1 AK401140 BAM17762.1 KPJ05764.1 AB260118 BAF91713.1 KF487630 AII01028.1 OWR47214.1 KF487614 AII01012.1 KX585429 ARO70322.1 AY701858 AAW23971.1 MG788191 AXF48709.1 JTDY01000771 KOB75936.1 KM365202 AIX97837.1 BABH01021426 DQ855517 ABH88204.1 KF026058 AHX37227.1 ODYU01000014 SOQ34069.1 AJ973465 CAJ01512.1 RBVL01000221 RKO10462.1 LC027714 BAV56817.1 EZ407257 ACX53813.1 KT357404 AND82452.1 GDQN01008076 JAT82978.1 HQ874662 AEX07267.1 PCG73969.1 KX655948 AOG12897.1 KT282978 ALS03835.1 KC507182 AGI37365.1 KT991358 ANA75024.1 KC907755 AGY49268.1 JX305305 AFR92094.1 PZC77877.1 ODYU01001383 SOQ37483.1 GCVX01000015 JAI18215.1 MH027981 AZB49399.1 GCVX01000010 JAI18220.1 KC907753 AGY49266.1 BABH01021467 AB243752 AJ973408 BAF34357.1 CAJ01455.1 KP975095 ALT31614.1 KR003783 ALC79597.1 KP975097 ALT31616.1 KC907752 AGY49265.1 KF487638 AII01036.1 GENK01000040 JAV45873.1 MG546593 AYD42216.1 KF026052 AHX37221.1 KT261672 ALJ30212.1 KT381691 AMA98182.1 MG788201 AXF48719.1 KF487622 AII01020.1 MH027979 AZB49397.1 PCG73975.1 LC027706 BAV56809.1 EZ407157 ACX53719.1 KP729029 AKT26487.1 KC907749 AGY49262.1 MG788187 AXF48705.1 APGK01034971 KC113424 KB740923 AGI05172.1 ENN78117.1 KQ414578 KOC71174.1 AGBW02009338 OWR51069.1 AF481963 DQ855484 AAN59784.1 ABH88171.1 MF770646 AXG21599.1 PZC77872.1 KX585420 ARO70313.1 KP729030 AKT26488.1 AJ973458 CAJ01505.1 KU133770 ALV87595.1
AOG12890.1 LC027703 BAV56806.1 MG788188 AXF48706.1 MH027976 AZB49394.1 HQ649755 ADV36661.1 KF026051 AHX37220.1 KT357396 AND82444.1 KX585414 ARO70307.1 KF487616 AII01014.1 KT282984 ALS03841.1 GENK01000048 JAV45865.1 KT991357 ANA75023.1 KP729031 AKT26489.1 KT261677 ALJ30217.1 GDQN01007391 GDQN01007068 JAT83663.1 JAT83986.1 KC507180 AGI37363.1 KX954372 ATD12156.1 GAIX01008045 JAA84515.1 JX863697 AGR39572.1 AB260121 BAF91716.1 KQ458575 KPJ05768.1 KY815027 ASA40088.1 KZ149917 PZC77881.1 KF487636 AII01034.1 KM236063 AIW65100.1 AGBW02011101 OWR47215.1 KQ461073 KPJ09662.1 MH050646 AZT78915.1 NWSH01000827 PCG73980.1 MG546594 AYD42217.1 JTDY01001439 KOB73859.1 MG788186 AXF48704.1 AJ973462 CAJ01509.1 EF202829 FJ948817 ABM92664.1 ACR43877.1 KPJ09666.1 AGBW02009828 OWR49808.1 AK401140 BAM17762.1 KPJ05764.1 AB260118 BAF91713.1 KF487630 AII01028.1 OWR47214.1 KF487614 AII01012.1 KX585429 ARO70322.1 AY701858 AAW23971.1 MG788191 AXF48709.1 JTDY01000771 KOB75936.1 KM365202 AIX97837.1 BABH01021426 DQ855517 ABH88204.1 KF026058 AHX37227.1 ODYU01000014 SOQ34069.1 AJ973465 CAJ01512.1 RBVL01000221 RKO10462.1 LC027714 BAV56817.1 EZ407257 ACX53813.1 KT357404 AND82452.1 GDQN01008076 JAT82978.1 HQ874662 AEX07267.1 PCG73969.1 KX655948 AOG12897.1 KT282978 ALS03835.1 KC507182 AGI37365.1 KT991358 ANA75024.1 KC907755 AGY49268.1 JX305305 AFR92094.1 PZC77877.1 ODYU01001383 SOQ37483.1 GCVX01000015 JAI18215.1 MH027981 AZB49399.1 GCVX01000010 JAI18220.1 KC907753 AGY49266.1 BABH01021467 AB243752 AJ973408 BAF34357.1 CAJ01455.1 KP975095 ALT31614.1 KR003783 ALC79597.1 KP975097 ALT31616.1 KC907752 AGY49265.1 KF487638 AII01036.1 GENK01000040 JAV45873.1 MG546593 AYD42216.1 KF026052 AHX37221.1 KT261672 ALJ30212.1 KT381691 AMA98182.1 MG788201 AXF48719.1 KF487622 AII01020.1 MH027979 AZB49397.1 PCG73975.1 LC027706 BAV56809.1 EZ407157 ACX53719.1 KP729029 AKT26487.1 KC907749 AGY49262.1 MG788187 AXF48705.1 APGK01034971 KC113424 KB740923 AGI05172.1 ENN78117.1 KQ414578 KOC71174.1 AGBW02009338 OWR51069.1 AF481963 DQ855484 AAN59784.1 ABH88171.1 MF770646 AXG21599.1 PZC77872.1 KX585420 ARO70313.1 KP729030 AKT26488.1 AJ973458 CAJ01505.1 KU133770 ALV87595.1
Proteomes
Pfam
PF03392 OS-D
SUPFAM
SSF100910
SSF100910
Gene 3D
ProteinModelPortal
Q3LB97
A0A1B3P5K5
A0A1B4ZBJ1
A0A345BEN2
A0A3G6V6M0
E7EC45
+ More
A0A0C4KD22 A0A172QDN4 A0A2K8GKZ4 A0A076E960 A0A1L2BLB2 A0A1V1WBX1 A0A1W5LB26 A0A0K1DCX9 A0A0P0EW15 A0A1E1W9K9 M4XYS2 A0A290U466 S4PC20 U5KD25 A8QWQ6 A0A194QJL9 A0A1Z2R8Q9 A0A2W1BU58 A0A076E970 A0A0A0VG76 A0A212F0D7 A0A194QW11 A0A3Q9NAY6 A0A2A4JRD7 A0A386H8V2 A0A0L7LEQ2 A0A345BEN0 Q3LB44 A2TIK5 A0A194R1E1 A0A212F7W2 I4DIM2 A0A194QK61 A8QWQ3 A0A076E964 A0A212F0I9 A0A076E7E7 A0A2K8GL22 Q4VR77 A0A345BEN5 A0A0L7LL43 A0A0A1CR78 Q0MRJ2 A0A0C4KCE8 A0A2H1UZS0 Q3LB41 A0A3B0EM79 A0A1B4ZBL4 D2SNX2 A0A172QDP1 A0A1E1W7L4 H6TDX9 A0A2A4JQ79 A0A1B3P5L7 A0A1L2BL66 M4Y081 A0A1W5LAR7 U5PZS2 J9VHK5 A0A2H1V9F0 A0A0K8TVZ7 A0A3G6V6Z3 A0A0K8TW01 U5PZD9 Q3LB98 A0A0U2SQP9 A0A0M4FCQ3 A0A0U3AMZ7 U5Q2S2 A0A076E5V2 A0A1V1WC03 A0A386H9C5 A0A0C4K658 A0A0P0EM27 A0A109ZT31 A0A345BEP5 A0A076ED87 A0A3G6V6L7 A0A2A4JPI3 A0A1B4ZBL1 D2SNM8 A0A0K1DDI7 U5PZ61 A0A345BEN1 M4VTH9 A0A0L7RK18 A0A212FBG3 Q8I6X7 A0A345F0U4 A0A2W1BS20 A0A2K8GL19 A0A0K1DDT3 Q3LB48 A0A0U3T5N0
A0A0C4KD22 A0A172QDN4 A0A2K8GKZ4 A0A076E960 A0A1L2BLB2 A0A1V1WBX1 A0A1W5LB26 A0A0K1DCX9 A0A0P0EW15 A0A1E1W9K9 M4XYS2 A0A290U466 S4PC20 U5KD25 A8QWQ6 A0A194QJL9 A0A1Z2R8Q9 A0A2W1BU58 A0A076E970 A0A0A0VG76 A0A212F0D7 A0A194QW11 A0A3Q9NAY6 A0A2A4JRD7 A0A386H8V2 A0A0L7LEQ2 A0A345BEN0 Q3LB44 A2TIK5 A0A194R1E1 A0A212F7W2 I4DIM2 A0A194QK61 A8QWQ3 A0A076E964 A0A212F0I9 A0A076E7E7 A0A2K8GL22 Q4VR77 A0A345BEN5 A0A0L7LL43 A0A0A1CR78 Q0MRJ2 A0A0C4KCE8 A0A2H1UZS0 Q3LB41 A0A3B0EM79 A0A1B4ZBL4 D2SNX2 A0A172QDP1 A0A1E1W7L4 H6TDX9 A0A2A4JQ79 A0A1B3P5L7 A0A1L2BL66 M4Y081 A0A1W5LAR7 U5PZS2 J9VHK5 A0A2H1V9F0 A0A0K8TVZ7 A0A3G6V6Z3 A0A0K8TW01 U5PZD9 Q3LB98 A0A0U2SQP9 A0A0M4FCQ3 A0A0U3AMZ7 U5Q2S2 A0A076E5V2 A0A1V1WC03 A0A386H9C5 A0A0C4K658 A0A0P0EM27 A0A109ZT31 A0A345BEP5 A0A076ED87 A0A3G6V6L7 A0A2A4JPI3 A0A1B4ZBL1 D2SNM8 A0A0K1DDI7 U5PZ61 A0A345BEN1 M4VTH9 A0A0L7RK18 A0A212FBG3 Q8I6X7 A0A345F0U4 A0A2W1BS20 A0A2K8GL19 A0A0K1DDT3 Q3LB48 A0A0U3T5N0
PDB
2GVS
E-value=7.09984e-18,
Score=215
Ontologies
GO
PANTHER
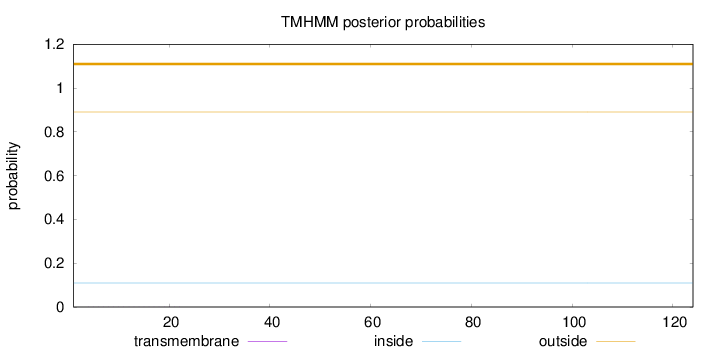
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.996976
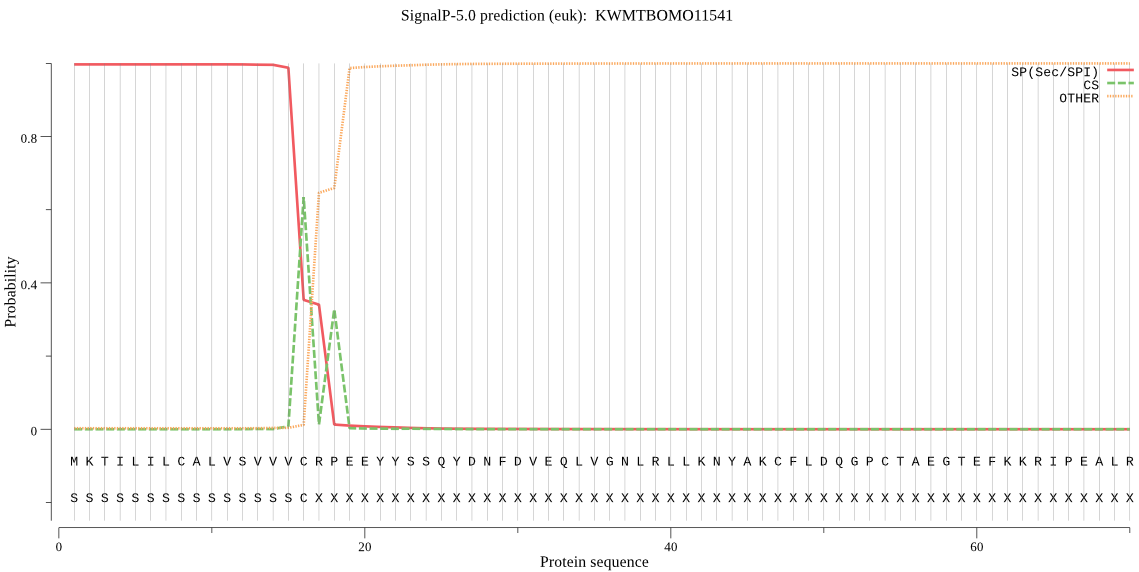
Length:
124
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00493
Exp number, first 60 AAs:
0.00493
Total prob of N-in:
0.10963
outside
1 - 124
Population Genetic Test Statistics
Pi
189.494701
Theta
162.046788
Tajima's D
-0.839917
CLR
0.750243
CSRT
0.172691365431728
Interpretation
Uncertain
