Gene
KWMTBOMO11523
Pre Gene Modal
BGIBMGA004062
Annotation
PREDICTED:_putative_glycerol_kinase_5_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.26
Sequence
CDS
ATGGAAGAGAAATATATTTTAACTTTAGATATAGGAACTACGACAATTCGTGCGTTTATTTATAATTCTAGAGCAGAAACTGTAGGAAAAGCTGTCGATCAGGTGATTCTGCACTATCCCTGTCCTGGATTTGTGGAGATTAACCCAAATGAATTATGGACCTCAGTTATTTCAGTGGTGAAGAAATCATTTCAAAATGCCAATCTTTCGGCTGGTCAGATCACAGCAATGGGTATATCTACTCAAAGGAGCACATTCATCACGTGGTCTCGTACAACTGGCAAGCATTTTCACAATTTTGTAACTTGGAAAGACTTAAGAGCTGACAGTCTTGTCAAACAGTGGAACAAGTCTTATACATGGAAGTTATTTAAATTCAGCGCCTACGTTCTATATTTGGTGTCACGGAGCAAAAGATTCAAAGCCGGAAGTGTATTCAAATTTATGAATACGCAGACCTCGCTGAGATTGAATTGGGTTCTTCAAAATGTAGTTGCGTTAAGAAACGCTGTACAAAACAACGACGCAATGTTTGGAACCTTGGACACGTGGTTATTATATAAATTGACAGGTCAACGTATTCACATGACCGATGTTTCTTCGGCATCCGCTACAGGCTTCTACGATCCGTTTACGATGCAGTGGGCCGGCTGGGGAACACTCTTATTAAATATTCCGATTTCCTCTTTGCCGACGGTCGTGGACACGGCCGGCGAACACTTCACCAGCACGGCGCCGGATATATGGGGTCATGCCATTCCGATCAGAAGCTGCATGGCCGATCAAACGGCGTCGATGTGGGGCTCGTGTTGTTTTGACGTTGGTGACATCAAACTGACAATGGGAACGGGGACGTTCTTTAATATAAACACCGGATCGGCACCTCACGCGAGCGTGTCCGGCCTGTACCCGGTGGTCGGCTGGAGGCTCGGCGACGAACTGGTGTATTCCGCGGAAGGAGCGAACAACGATACGGCTTCTATAATTAAATGGGCACAAAATTTAGGTTTATTTATCAGTCCGCAAGAGACTGCGGACATAGCTAATTCAGTAACGGATACGGATGGCGCCTTCTTCATTCCCGCTTTTAGTGGATTAGGGCCCCCATATAACGACGGTACAGCCGCTTCAGGTTTCATAGGCATGAAGCCATCGACAACAAAAGCGCATTTGGTTCGCGCGTTACTAGAGTCATTAGCGTTTCGTACCGCGCAATTATATCAGTGTGTGCGTAATGAAACCGATTACACGTTTCATTCTATAAGGCTAGACGGTGGTGTGTCTAACAATGATTTCCTCGCCCAACTGGTTTCGGACTTAACGGGTCTACGCGTGGAGAGACCCGTGCACGTAGAGATGTCCTCTTTAGGATGCGCGCATATTGTTGGACTACATCTAGGTATATGGAAATCGAAACAGGAACTAAAATCTCTAAGGAAAGTGGGATGCGTGTTTCATCCACGGCCGCACGTGAAAAAATCTTACGAACACACGATCTCGATGTGGGAAGACGCCGTCACACGGATGTGCGGATGGTATTCAAGAAGCAATTCGTTGAAGTCAGTCAACGCTGTATCGGACTCCAGTGGTTGTGGAGTAGTTCCTAAGAATAGCTTTAAACTGCTAAAGAATAACGGGGACGCAGTGAAATAA
Protein
MEEKYILTLDIGTTTIRAFIYNSRAETVGKAVDQVILHYPCPGFVEINPNELWTSVISVVKKSFQNANLSAGQITAMGISTQRSTFITWSRTTGKHFHNFVTWKDLRADSLVKQWNKSYTWKLFKFSAYVLYLVSRSKRFKAGSVFKFMNTQTSLRLNWVLQNVVALRNAVQNNDAMFGTLDTWLLYKLTGQRIHMTDVSSASATGFYDPFTMQWAGWGTLLLNIPISSLPTVVDTAGEHFTSTAPDIWGHAIPIRSCMADQTASMWGSCCFDVGDIKLTMGTGTFFNINTGSAPHASVSGLYPVVGWRLGDELVYSAEGANNDTASIIKWAQNLGLFISPQETADIANSVTDTDGAFFIPAFSGLGPPYNDGTAASGFIGMKPSTTKAHLVRALLESLAFRTAQLYQCVRNETDYTFHSIRLDGGVSNNDFLAQLVSDLTGLRVERPVHVEMSSLGCAHIVGLHLGIWKSKQELKSLRKVGCVFHPRPHVKKSYEHTISMWEDAVTRMCGWYSRSNSLKSVNAVSDSSGCGVVPKNSFKLLKNNGDAVK
Summary
Similarity
Belongs to the FGGY kinase family.
Belongs to the aldehyde dehydrogenase family.
Belongs to the aldehyde dehydrogenase family.
Uniprot
H9J3H5
A0A2H1WB10
A0A194QXK1
A0A2W1B9Y6
A0A067QVT9
A0A1B6D2A8
+ More
E0W2J6 A0A2J7RN04 A0A195FQ80 A0A1B6KQ55 A0A195ELQ9 A0A151XK53 A0A2J7RMY8 A0A1B6G912 A0A195B5N4 F4WCH8 A0A232FG77 A0A158NIZ3 K7J3Y8 E9IUI8 A0A1B6DGB8 A0A026WSF8 A0A0L7QKF3 T1I0C8 E2A121 A0A023EZE0 A0A2M4BK00 W5MMV1 A0A084WF05 A0A2K6VBA6 A0A088ANW9 A0A1S4FJK7 A0A182XZ46 A0A240PKQ7 A0A1I8JSQ6 A0A182GPH7 A0A182UW30 A0A2M4AQH1 A0A1I8JVW5 A0A182IS65 A0A182TNN0 A0A240PPN5 Q7QJM6 A0A1Y9GLC2 A0A1Q3FNA3 A0A182KRN8 A0A182GES2 H3AHT7 A0A1Y9IW38 A0A182RQ04 A0A1Y1L423 A0A182M0E2 A0A1L8E1V6 A0A1Y9H2M2 A0A2I4CH18 A0A3P8YJD2 A0A182QRJ9 A0A1L8E237 A0A1L8E1T9 A0A1S3QYK3 A0A3P8WKJ1 A0A3P9A5S9 A0A1W4YUQ8 A0A146NSJ4 A0A3B4AJ38 B0VZT8 A0A3Q3A057 D6WU91 A0A3B4Z0K2 A0A154PQI6 A0A3Q2PRH5 A0A3B4ERW9 F1QS50 W5J793 A0A3B4XWN7 C3ZEB8 A0A0S7M188 A0A1A8MSW1 A0A3P8WNZ0 A0A3P9BAB8 A0A146N557 I3JWW7 A0A2K6S3X9 A0A3B3Y6Z1 A0A3B5QSC8 A0A087YFV2 V4A8I4 A0A340WJ25 A0A3B3TQ25 V9KCQ3 A0A3Q1G047 A0A2K5ZB75 A0A2K5M4M3 F7F132 H3CQX6 A0A1A8AC09 A0A3Q4G8U4 A0A2K6NYM2 A0A096MAN5
E0W2J6 A0A2J7RN04 A0A195FQ80 A0A1B6KQ55 A0A195ELQ9 A0A151XK53 A0A2J7RMY8 A0A1B6G912 A0A195B5N4 F4WCH8 A0A232FG77 A0A158NIZ3 K7J3Y8 E9IUI8 A0A1B6DGB8 A0A026WSF8 A0A0L7QKF3 T1I0C8 E2A121 A0A023EZE0 A0A2M4BK00 W5MMV1 A0A084WF05 A0A2K6VBA6 A0A088ANW9 A0A1S4FJK7 A0A182XZ46 A0A240PKQ7 A0A1I8JSQ6 A0A182GPH7 A0A182UW30 A0A2M4AQH1 A0A1I8JVW5 A0A182IS65 A0A182TNN0 A0A240PPN5 Q7QJM6 A0A1Y9GLC2 A0A1Q3FNA3 A0A182KRN8 A0A182GES2 H3AHT7 A0A1Y9IW38 A0A182RQ04 A0A1Y1L423 A0A182M0E2 A0A1L8E1V6 A0A1Y9H2M2 A0A2I4CH18 A0A3P8YJD2 A0A182QRJ9 A0A1L8E237 A0A1L8E1T9 A0A1S3QYK3 A0A3P8WKJ1 A0A3P9A5S9 A0A1W4YUQ8 A0A146NSJ4 A0A3B4AJ38 B0VZT8 A0A3Q3A057 D6WU91 A0A3B4Z0K2 A0A154PQI6 A0A3Q2PRH5 A0A3B4ERW9 F1QS50 W5J793 A0A3B4XWN7 C3ZEB8 A0A0S7M188 A0A1A8MSW1 A0A3P8WNZ0 A0A3P9BAB8 A0A146N557 I3JWW7 A0A2K6S3X9 A0A3B3Y6Z1 A0A3B5QSC8 A0A087YFV2 V4A8I4 A0A340WJ25 A0A3B3TQ25 V9KCQ3 A0A3Q1G047 A0A2K5ZB75 A0A2K5M4M3 F7F132 H3CQX6 A0A1A8AC09 A0A3Q4G8U4 A0A2K6NYM2 A0A096MAN5
Pubmed
19121390
26354079
28756777
24845553
20566863
21719571
+ More
28648823 21347285 20075255 21282665 24508170 30249741 20798317 25474469 24438588 20920257 25244985 26483478 12364791 14747013 17210077 20966253 9215903 28004739 25069045 24487278 25463417 18362917 19820115 23594743 18563158 25186727 23542700 23254933 24402279 25243066 15496914 25362486
28648823 21347285 20075255 21282665 24508170 30249741 20798317 25474469 24438588 20920257 25244985 26483478 12364791 14747013 17210077 20966253 9215903 28004739 25069045 24487278 25463417 18362917 19820115 23594743 18563158 25186727 23542700 23254933 24402279 25243066 15496914 25362486
EMBL
BABH01021416
ODYU01007459
SOQ50248.1
KQ461073
KPJ09685.1
KZ150235
+ More
PZC71968.1 KK852954 KDR13305.1 GEDC01017471 JAS19827.1 DS235878 EEB19852.1 NEVH01002546 PNF42202.1 KQ981305 KYN42745.1 GEBQ01026440 JAT13537.1 KQ978739 KYN28862.1 KQ982052 KYQ60711.1 PNF42203.1 GECZ01011006 JAS58763.1 KQ976582 KYM79828.1 GL888070 EGI68158.1 NNAY01000288 OXU29458.1 ADTU01001872 ADTU01001873 ADTU01001874 AAZX01001002 GL765979 EFZ15753.1 GEDC01012600 JAS24698.1 KK107119 QOIP01000009 EZA58591.1 RLU19133.1 KQ414986 KOC59001.1 ACPB03020901 GL435638 EFN72874.1 GBBI01004114 JAC14598.1 GGFJ01004193 MBW53334.1 AHAT01015288 AHAT01015289 ATLV01023269 KE525341 KFB48799.1 GGFL01005915 MBW70093.1 JXUM01014982 JXUM01014983 JXUM01014984 KQ560418 KXJ82509.1 GGFK01009641 MBW42962.1 AAAB01008807 EAA04771.4 APCN01000213 GFDL01006053 JAV28992.1 JXUM01058605 KQ562010 KXJ76915.1 AFYH01135225 AFYH01135226 AFYH01135227 AFYH01135228 AFYH01135229 GEZM01071387 JAV65827.1 AXCM01000276 GFDF01001383 JAV12701.1 AXCN02000894 GFDF01001385 JAV12699.1 GFDF01001384 JAV12700.1 GCES01152176 JAQ34146.1 DS231815 EDS35180.1 KQ971352 EFA06784.1 KQ435007 KZC13518.1 CU929296 CU929319 ADMH02002032 ETN59846.1 GG666612 EEN49074.1 GBYX01128092 JAO95390.1 HAEF01018512 SBR59671.1 GCES01159331 JAQ26991.1 AERX01001365 AYCK01012698 KB200786 ESP00284.1 JW862984 AFO95501.1 GAMT01008495 GAMS01009281 GAMR01005403 JAB03366.1 JAB13855.1 JAB28529.1 HADY01013275 HAEJ01013163 SBP51760.1
PZC71968.1 KK852954 KDR13305.1 GEDC01017471 JAS19827.1 DS235878 EEB19852.1 NEVH01002546 PNF42202.1 KQ981305 KYN42745.1 GEBQ01026440 JAT13537.1 KQ978739 KYN28862.1 KQ982052 KYQ60711.1 PNF42203.1 GECZ01011006 JAS58763.1 KQ976582 KYM79828.1 GL888070 EGI68158.1 NNAY01000288 OXU29458.1 ADTU01001872 ADTU01001873 ADTU01001874 AAZX01001002 GL765979 EFZ15753.1 GEDC01012600 JAS24698.1 KK107119 QOIP01000009 EZA58591.1 RLU19133.1 KQ414986 KOC59001.1 ACPB03020901 GL435638 EFN72874.1 GBBI01004114 JAC14598.1 GGFJ01004193 MBW53334.1 AHAT01015288 AHAT01015289 ATLV01023269 KE525341 KFB48799.1 GGFL01005915 MBW70093.1 JXUM01014982 JXUM01014983 JXUM01014984 KQ560418 KXJ82509.1 GGFK01009641 MBW42962.1 AAAB01008807 EAA04771.4 APCN01000213 GFDL01006053 JAV28992.1 JXUM01058605 KQ562010 KXJ76915.1 AFYH01135225 AFYH01135226 AFYH01135227 AFYH01135228 AFYH01135229 GEZM01071387 JAV65827.1 AXCM01000276 GFDF01001383 JAV12701.1 AXCN02000894 GFDF01001385 JAV12699.1 GFDF01001384 JAV12700.1 GCES01152176 JAQ34146.1 DS231815 EDS35180.1 KQ971352 EFA06784.1 KQ435007 KZC13518.1 CU929296 CU929319 ADMH02002032 ETN59846.1 GG666612 EEN49074.1 GBYX01128092 JAO95390.1 HAEF01018512 SBR59671.1 GCES01159331 JAQ26991.1 AERX01001365 AYCK01012698 KB200786 ESP00284.1 JW862984 AFO95501.1 GAMT01008495 GAMS01009281 GAMR01005403 JAB03366.1 JAB13855.1 JAB28529.1 HADY01013275 HAEJ01013163 SBP51760.1
Proteomes
UP000005204
UP000053240
UP000027135
UP000009046
UP000235965
UP000078541
+ More
UP000078492 UP000075809 UP000078540 UP000007755 UP000215335 UP000005205 UP000002358 UP000053097 UP000279307 UP000053825 UP000015103 UP000000311 UP000018468 UP000030765 UP000005203 UP000076408 UP000075881 UP000069272 UP000069940 UP000249989 UP000075903 UP000076407 UP000075880 UP000075902 UP000075885 UP000007062 UP000075840 UP000075882 UP000008672 UP000075920 UP000075900 UP000075883 UP000075884 UP000192220 UP000265140 UP000075886 UP000087266 UP000265120 UP000192224 UP000261520 UP000002320 UP000264800 UP000007266 UP000261400 UP000076502 UP000265000 UP000261460 UP000000437 UP000000673 UP000261360 UP000001554 UP000265160 UP000005207 UP000233220 UP000261480 UP000002852 UP000028760 UP000030746 UP000265300 UP000261500 UP000257200 UP000233140 UP000233060 UP000008225 UP000007303 UP000261580 UP000233200
UP000078492 UP000075809 UP000078540 UP000007755 UP000215335 UP000005205 UP000002358 UP000053097 UP000279307 UP000053825 UP000015103 UP000000311 UP000018468 UP000030765 UP000005203 UP000076408 UP000075881 UP000069272 UP000069940 UP000249989 UP000075903 UP000076407 UP000075880 UP000075902 UP000075885 UP000007062 UP000075840 UP000075882 UP000008672 UP000075920 UP000075900 UP000075883 UP000075884 UP000192220 UP000265140 UP000075886 UP000087266 UP000265120 UP000192224 UP000261520 UP000002320 UP000264800 UP000007266 UP000261400 UP000076502 UP000265000 UP000261460 UP000000437 UP000000673 UP000261360 UP000001554 UP000265160 UP000005207 UP000233220 UP000261480 UP000002852 UP000028760 UP000030746 UP000265300 UP000261500 UP000257200 UP000233140 UP000233060 UP000008225 UP000007303 UP000261580 UP000233200
PRIDE
Interpro
IPR000577
Carb_kinase_FGGY
+ More
IPR018483 Carb_kinase_FGGY_CS
IPR018484 Carb_kinase_FGGY_N
IPR018485 Carb_kinase_FGGY_C
IPR037444 GK5_FGGY
IPR021131 Ribosomal_L18e/L15P
IPR036227 L18e/L15P_sf
IPR015590 Aldehyde_DH_dom
IPR029510 Ald_DH_CS_GLU
IPR016162 Ald_DH_N
IPR016163 Ald_DH_C
IPR016161 Ald_DH/histidinol_DH
IPR021132 Ribosomal_L18e_CS
IPR018483 Carb_kinase_FGGY_CS
IPR018484 Carb_kinase_FGGY_N
IPR018485 Carb_kinase_FGGY_C
IPR037444 GK5_FGGY
IPR021131 Ribosomal_L18e/L15P
IPR036227 L18e/L15P_sf
IPR015590 Aldehyde_DH_dom
IPR029510 Ald_DH_CS_GLU
IPR016162 Ald_DH_N
IPR016163 Ald_DH_C
IPR016161 Ald_DH/histidinol_DH
IPR021132 Ribosomal_L18e_CS
Gene 3D
ProteinModelPortal
H9J3H5
A0A2H1WB10
A0A194QXK1
A0A2W1B9Y6
A0A067QVT9
A0A1B6D2A8
+ More
E0W2J6 A0A2J7RN04 A0A195FQ80 A0A1B6KQ55 A0A195ELQ9 A0A151XK53 A0A2J7RMY8 A0A1B6G912 A0A195B5N4 F4WCH8 A0A232FG77 A0A158NIZ3 K7J3Y8 E9IUI8 A0A1B6DGB8 A0A026WSF8 A0A0L7QKF3 T1I0C8 E2A121 A0A023EZE0 A0A2M4BK00 W5MMV1 A0A084WF05 A0A2K6VBA6 A0A088ANW9 A0A1S4FJK7 A0A182XZ46 A0A240PKQ7 A0A1I8JSQ6 A0A182GPH7 A0A182UW30 A0A2M4AQH1 A0A1I8JVW5 A0A182IS65 A0A182TNN0 A0A240PPN5 Q7QJM6 A0A1Y9GLC2 A0A1Q3FNA3 A0A182KRN8 A0A182GES2 H3AHT7 A0A1Y9IW38 A0A182RQ04 A0A1Y1L423 A0A182M0E2 A0A1L8E1V6 A0A1Y9H2M2 A0A2I4CH18 A0A3P8YJD2 A0A182QRJ9 A0A1L8E237 A0A1L8E1T9 A0A1S3QYK3 A0A3P8WKJ1 A0A3P9A5S9 A0A1W4YUQ8 A0A146NSJ4 A0A3B4AJ38 B0VZT8 A0A3Q3A057 D6WU91 A0A3B4Z0K2 A0A154PQI6 A0A3Q2PRH5 A0A3B4ERW9 F1QS50 W5J793 A0A3B4XWN7 C3ZEB8 A0A0S7M188 A0A1A8MSW1 A0A3P8WNZ0 A0A3P9BAB8 A0A146N557 I3JWW7 A0A2K6S3X9 A0A3B3Y6Z1 A0A3B5QSC8 A0A087YFV2 V4A8I4 A0A340WJ25 A0A3B3TQ25 V9KCQ3 A0A3Q1G047 A0A2K5ZB75 A0A2K5M4M3 F7F132 H3CQX6 A0A1A8AC09 A0A3Q4G8U4 A0A2K6NYM2 A0A096MAN5
E0W2J6 A0A2J7RN04 A0A195FQ80 A0A1B6KQ55 A0A195ELQ9 A0A151XK53 A0A2J7RMY8 A0A1B6G912 A0A195B5N4 F4WCH8 A0A232FG77 A0A158NIZ3 K7J3Y8 E9IUI8 A0A1B6DGB8 A0A026WSF8 A0A0L7QKF3 T1I0C8 E2A121 A0A023EZE0 A0A2M4BK00 W5MMV1 A0A084WF05 A0A2K6VBA6 A0A088ANW9 A0A1S4FJK7 A0A182XZ46 A0A240PKQ7 A0A1I8JSQ6 A0A182GPH7 A0A182UW30 A0A2M4AQH1 A0A1I8JVW5 A0A182IS65 A0A182TNN0 A0A240PPN5 Q7QJM6 A0A1Y9GLC2 A0A1Q3FNA3 A0A182KRN8 A0A182GES2 H3AHT7 A0A1Y9IW38 A0A182RQ04 A0A1Y1L423 A0A182M0E2 A0A1L8E1V6 A0A1Y9H2M2 A0A2I4CH18 A0A3P8YJD2 A0A182QRJ9 A0A1L8E237 A0A1L8E1T9 A0A1S3QYK3 A0A3P8WKJ1 A0A3P9A5S9 A0A1W4YUQ8 A0A146NSJ4 A0A3B4AJ38 B0VZT8 A0A3Q3A057 D6WU91 A0A3B4Z0K2 A0A154PQI6 A0A3Q2PRH5 A0A3B4ERW9 F1QS50 W5J793 A0A3B4XWN7 C3ZEB8 A0A0S7M188 A0A1A8MSW1 A0A3P8WNZ0 A0A3P9BAB8 A0A146N557 I3JWW7 A0A2K6S3X9 A0A3B3Y6Z1 A0A3B5QSC8 A0A087YFV2 V4A8I4 A0A340WJ25 A0A3B3TQ25 V9KCQ3 A0A3Q1G047 A0A2K5ZB75 A0A2K5M4M3 F7F132 H3CQX6 A0A1A8AC09 A0A3Q4G8U4 A0A2K6NYM2 A0A096MAN5
PDB
3GE1
E-value=1.7513e-78,
Score=746
Ontologies
KEGG
GO
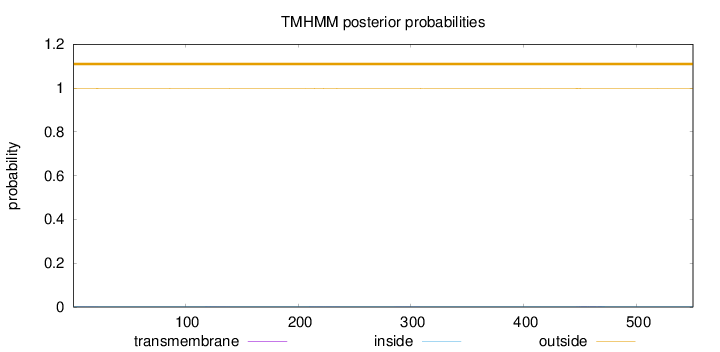
Topology
Length:
550
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0872600000000001
Exp number, first 60 AAs:
0.00988999999999999
Total prob of N-in:
0.00293
outside
1 - 550
Population Genetic Test Statistics
Pi
167.785635
Theta
155.712119
Tajima's D
-0.887805
CLR
142.484936
CSRT
0.156242187890605
Interpretation
Uncertain
