Gene
KWMTBOMO11518
Annotation
reverse_transcriptase_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.622 Nuclear Reliability : 1.145
Sequence
CDS
ATGACCGCTACCAAGAGGCAGCAGGCACTCGTGGGGCCACGGGAGCTGGCTGGAAACTTCAGCCTGGACGAGGTCATCACTCGCACCGTCCAGGTTGTATTGGAAAGGCTGGACGGCAGGCTGGCGGCCCTCGAGGCGCAACTCATTGCCCCGGCTGACGCACGGCGACCGGCACATGCAGCAGTGTTGGGTGCTCCAGCCGCCCACCCCGCCCCCGTACAGACTCCAGCCACTAGGTTGGGCCCGGGACGGGATAGAACAAAGCGGGGGGGCGGCGCCAACGCGAAACGCCCATCGCAAGCACCCCAGCCCCGTCCGCTGCCTCCGCCCCCATCCAACATGGACGAGGGGTGGAACGTGGTGGCCCGGCGGGGTGCCAAGAAGGTCCCACTGGCAGCTCCACAAGTGGGTGAAGCGCCGGCGGCCACGGCGGCTCAGGCTGCTCCCCAACAGGGTCGAGCGCCTGTAGCCGCCAAAAAGAAAAACAAGAAAACAAAGAAGCCACCGCGCGCGCCGCGGTCGGCGGCAGTGGTCCTAACGCTGCTGCCGGCTGCCAAAGCCAAGGGCATGACATACGGAGATGTCATTGCTCGGGCGCGCGCGGCAGTCGATCTGAATGAGATCGGGGCGGGGGAGGGTCTCCGCTCTAGGCAGACAGCCAATGGGGCCAAACTGCTAGAGTGTTCCGGTGCTGATAGTCCCGGCATCGCAGAGCGTCTGGCCGCAAGGCTCCGCGCGGTCCTGCCTGAAGAGGAGGTGCGGGTGCATAGGCCGGTCAAGATGGCCGAATTGAAAGTGACCGGCCTGGACGAATGCACCACCAGGGATGAGGTGGTGGCGGCCGTCGCTTCCCAAGGCAACTGTGCCCTGCAGAATATAACCGTAGGCGAGCTGCGTCCCGGTTATAGCGGGGCCAGCTCTGTGTGGGTACGTTGCCCGGTGGAGGCGGCCGTCGCCCTGGCTGCTCCTCCGCCCGGGAGACCTTTGGGGGATCCGGGTAGGTTGCGCGTGGGCTGGATAGCAGCCCACGTGCGCCTGCTAGAACCACGCGCCTGGCGGTGCCTAAAGTGCTTCAGCACTGGGCACTGCCTCGCCAGGTGCGTAAGTGCGGTTGACCGCAGCGGACTGTGTTTCCGCTGTGGTCAGCCTGGACACAAGGCGGCCGTGTGCTCGGCTGCGCCGCATTGCTTATTGTGTGCGGCGGCCGGACGCAGGGCTGACCACCGGACCGGGGGCAAGGCATGCCCCCCAGCCGGCAAAAACGCAATCAGAAATTGGCGCCGCCAAGCTAAGCGGCGCCAAAAGAAGCTGTCGGCCGGGGGAGCACCGGCCGGCACTTTGGCCCCCGCTGAGGCGTTCGCGCCCGATAGCGGGGACAATGGAAAGATCCGCGATGATTCCCCCCTGGCGATGGTGACCAGGGGTCCCGGGATCGTCGCGGTACAATTAGAAAATACCGTCGTGATCGGGGTGTACTTCTCTCCGAATCGGCCTACCGTCGAGTTCGAGCGGTTCCTGGACGGGCTAGAGGTGATCGCTCGTCACCTCTCTCCCCGTTCCGTGATACTGGCGGGGGACTTCAATGCGAAGTCTGTCTCTTGGGGTTCCCCATCTACGGACGCTCGTGGTAGGCTGTTGGAGGAGTGGGCGGTCGCGGCCGATCTCTGTATCGTTAACAGGGGCTCAGTCGCGACATGCGTGCGGTGGACAGGAGAGTCTATTGTGGACTTGACGTTCGCGAGCTCGTCCGTCGCACAGCGCATCCTCGGCTGGGGCGTGGTGGTGGGGGCGGAATCGTTGTCAGATCACCGGTATATCCGGTTCGATCTTTCCGCTCCCACGCCAACACCAGCCGCAGGCGCTCGCGGGGTTAACCCCCCGCGGAGTGCATCCCGGTCATTCCCTAGGTGGGCATTGAAGCTCCTGAAGAAGGAGCTCCTGGTGGAGGCCTCTATGGTGGCGGCGTGGACGCCCACGCGCCCACACCCTTTCGACGTGGAGGCCGAGGCCGAATGGTTCCGCGGCGCGATGCACCGCATATGCGATGCCTCGATGCCCCGGGTCGGCTCTCGGCATTGCGGACGGCGGCAGTCTCCCTGGTGGTCGCCTGAAATTGCGAGACTGCGCGCGGTCTCCATTCGGGCGAGCCGCCAATGCGCTAGACACCGCCGTCGCCGCCACCTGCGGCGCGACGACCCCGTCGCGTTTGCGGAGGAGGAAACCCGGCTGCACGGCGAATATCGCGCTGCGAAGGACGCCCTGCGGCTGGCCATCAGGCGGGCCAAGGAGCAGAACATGGAGAGACTCTTGGAGGCGCTCGAAGCGGATCCATGGGGGAGCCCGTACAGGTTAGTGCGCGGCAAGCTGCGCCCGTGGGCCCCCCCTATGACTGAGCGACTCCAGCCTCAACAGCTGCGGGACATCGTTTCGGCGTTGTTCCCGCAGGAGCGGGAGGGTTTTGTTCCCCCCGCTATGGGTGAGCCGTCGGACGCCATCGACGGTGAAGCCCCTGCTGAGGTGCCCCGTATTACTGGGGCAGAGCTCCGTGTGGCGGTTGTAAGGATGACTGCAAAGGACACGGCCCCCGGCCCGGACGGAGTCCATGGCCGGGTGCTGGCCTTGGCCCTCGATGCCCTAGGGGACCGGCTCCTGGAGCTATTTAATAGTTGCCTGGAGTCGGGACGGTTCCCGTTGCTCTGGCGGACGGGCAGACTTGTGTTGTTGCGGAAGGAGGGGCGCCCGGTGGATACAGCTGCCGGGTATCGTCCTATCGTACTGCTGGACGAGGCGGGAAAATTGCTGGAGCGTATTCTGGCTGCCCGCATCGTTCAGCACCTGGTCGGGGTTGGACCTGACCTGTCGGCGGAGCAGTTCGGCTTCCGGGAGGGCCGTTCGACCGTGGATGCAATACTTCGGGTGCGGGCCCTCTCGGACGAGGCTGTCTCACGGGGTGGGGTGGCGCTGGCGGTGTCTCTTGACATCGCCAACGCATTTAACACCCTGCCCTGGTCCGTGATAGGGGGGGCACTGGAGAGGCATGGAGTGCCCCTCTACCTCCGCCGGCTGGTTGGCTCCTATTTGGAGGCCAGGTCGGTCGAATGTACCGGGTACGGTGGGACCCTTCACCGTTTTCCGGTCGTGCGGGGTGTTCCGCAGGGGTCGGTTCTCGGACCCCTCTTGTGGAACATCGGGTACGACTGGGTGCTGAGGGGTGCCCTCCTCCCGGGCCTCCGCGTTATTTGTTACGCGGACGACACGTTGGTCGTGGCCCGGGGGATGATTATAGAGAGGGTCGGGGTGCAGTTGAGGTACCTCGGCCTCGTGTTGGACAGCCGGTGGGCTTTTCGTGCTCACTTTGCGGAGCTGGTCCCCCGATTGATGAGGACAGCCGGTTCTTTGAGCCGACTGCTCCCGAATATCGGGGGGCCGGATCAGGCTGTGCGCCGCCTCTACGCAGGGGTGGTGCGATCGATGGCCCTGTACGGTGCGCCTGTATGGGCTGAGCCACGCAACCGGGCCGCTATGGCTCGGTTCCTGCGCCGGCCGCAGCGCACCGTTGCTATCAGGGTCATCCGTGGATATCGCACCGTCTCCTTCGAGGCGGCGTGTGTGTTGGCGGGGACGCCGCCATGGGAGCTGGAGGCGGAGTCGCTCGCTGCCGACTATCGGTGGCGCAGCGAGCTTCGTGCTCTGGGCGTGGTGCGTCCCTCCAAAAGTGAGCTGCGGGCGCGGAAGGCCCATTCTCGGCGGTCCGTGCTCGAATCGTGGTCGAGGCGGTTGGCCAACCCCACGTGGGGTCTACGGACCGTTGAGGCGGTTTACCCAGTCTTTGATGACTGGGTGAATCGTGGCCGAGGACGCCTCACCTTCCGTCTGGTGCAGGTGCTGACTGGGCACGGATGCTTCGGAAAGTATCTGCACCGGATAGGAGCTGAGCCGACGACGAGGTGCCACCATTGTGGACACGACCTGGACACGGCGGAGCATACGCTCGCTGTCTGCCCCGCTTGGGAGGTGCAGCGCCGTGTCCTGGTCGCAAAGATAGGACCGGACTTGTCGCTGCCTGGCGTCGTGGCGTCGATGCTTGGCGGTGACGAGTCCTGGAAGGCTATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGAGTGAGGCAAAGCTCTCCTCACTACGCAGAAACCCGCCGCCGCCGAGCAGGGGGTCGGGACCGGGGTCTCGTCCGTAACCCGGCCCCCTAA
Protein
MTATKRQQALVGPRELAGNFSLDEVITRTVQVVLERLDGRLAALEAQLIAPADARRPAHAAVLGAPAAHPAPVQTPATRLGPGRDRTKRGGGANAKRPSQAPQPRPLPPPPSNMDEGWNVVARRGAKKVPLAAPQVGEAPAATAAQAAPQQGRAPVAAKKKNKKTKKPPRAPRSAAVVLTLLPAAKAKGMTYGDVIARARAAVDLNEIGAGEGLRSRQTANGAKLLECSGADSPGIAERLAARLRAVLPEEEVRVHRPVKMAELKVTGLDECTTRDEVVAAVASQGNCALQNITVGELRPGYSGASSVWVRCPVEAAVALAAPPPGRPLGDPGRLRVGWIAAHVRLLEPRAWRCLKCFSTGHCLARCVSAVDRSGLCFRCGQPGHKAAVCSAAPHCLLCAAAGRRADHRTGGKACPPAGKNAIRNWRRQAKRRQKKLSAGGAPAGTLAPAEAFAPDSGDNGKIRDDSPLAMVTRGPGIVAVQLENTVVIGVYFSPNRPTVEFERFLDGLEVIARHLSPRSVILAGDFNAKSVSWGSPSTDARGRLLEEWAVAADLCIVNRGSVATCVRWTGESIVDLTFASSSVAQRILGWGVVVGAESLSDHRYIRFDLSAPTPTPAAGARGVNPPRSASRSFPRWALKLLKKELLVEASMVAAWTPTRPHPFDVEAEAEWFRGAMHRICDASMPRVGSRHCGRRQSPWWSPEIARLRAVSIRASRQCARHRRRRHLRRDDPVAFAEEETRLHGEYRAAKDALRLAIRRAKEQNMERLLEALEADPWGSPYRLVRGKLRPWAPPMTERLQPQQLRDIVSALFPQEREGFVPPAMGEPSDAIDGEAPAEVPRITGAELRVAVVRMTAKDTAPGPDGVHGRVLALALDALGDRLLELFNSCLESGRFPLLWRTGRLVLLRKEGRPVDTAAGYRPIVLLDEAGKLLERILAARIVQHLVGVGPDLSAEQFGFREGRSTVDAILRVRALSDEAVSRGGVALAVSLDIANAFNTLPWSVIGGALERHGVPLYLRRLVGSYLEARSVECTGYGGTLHRFPVVRGVPQGSVLGPLLWNIGYDWVLRGALLPGLRVICYADDTLVVARGMIIERVGVQLRYLGLVLDSRWAFRAHFAELVPRLMRTAGSLSRLLPNIGGPDQAVRRLYAGVVRSMALYGAPVWAEPRNRAAMARFLRRPQRTVAIRVIRGYRTVSFEAACVLAGTPPWELEAESLAADYRWRSELRALGVVRPSKSELRARKAHSRRSVLESWSRRLANPTWGLRTVEAVYPVFDDWVNRGRGRLTFRLVQVLTGHGCFGKYLHRIGAEPTTRCHHCGHDLDTAEHTLAVCPAWEVQRRVLVAKIGPDLSLPGVVASMLGGDESWKAMLDFCECTISQKEAAGRVRQSSPHYAETRRRRAGGRDRGLVRNPAP
Summary
Uniprot
Pubmed
EMBL
KZ149896
PZC78733.1
LBMM01004272
KMQ92618.1
LBMM01012412
KMQ86235.1
+ More
LBMM01005474 KMQ91485.1 LBMM01008278 KMQ89009.1 LBMM01003988 KMQ92922.1 RSAL01003504 RVE40273.1 LBMM01006977 KMQ90149.1 LBMM01002052 KMQ95336.1 ABLF02011183 ABLF02041884 LBMM01008445 KMQ88871.1 ABLF02018942 ABLF02041885 ABLF02013358 ABLF02013361 ABLF02054869 ABLF02041886 ABLF02011238 GGMR01019493 MBY32112.1 ABLF02012448 ABLF02012450 ABLF02012455 ABLF02012460 ABLF02023257 ABLF02011927 GFXV01003903 MBW15708.1 ABLF02027248 ABLF02018098 ABLF02030663 ABLF02041312 ABLF02020969
LBMM01005474 KMQ91485.1 LBMM01008278 KMQ89009.1 LBMM01003988 KMQ92922.1 RSAL01003504 RVE40273.1 LBMM01006977 KMQ90149.1 LBMM01002052 KMQ95336.1 ABLF02011183 ABLF02041884 LBMM01008445 KMQ88871.1 ABLF02018942 ABLF02041885 ABLF02013358 ABLF02013361 ABLF02054869 ABLF02041886 ABLF02011238 GGMR01019493 MBY32112.1 ABLF02012448 ABLF02012450 ABLF02012455 ABLF02012460 ABLF02023257 ABLF02011927 GFXV01003903 MBW15708.1 ABLF02027248 ABLF02018098 ABLF02030663 ABLF02041312 ABLF02020969
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR002123 Plipid/glycerol_acylTrfase
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR024079 MetalloPept_cat_dom_sf
IPR001590 Peptidase_M12B
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
IPR002123 Plipid/glycerol_acylTrfase
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR024079 MetalloPept_cat_dom_sf
IPR001590 Peptidase_M12B
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
Gene 3D
ProteinModelPortal
PDB
1WDU
E-value=1.68603e-09,
Score=155
Ontologies
GO
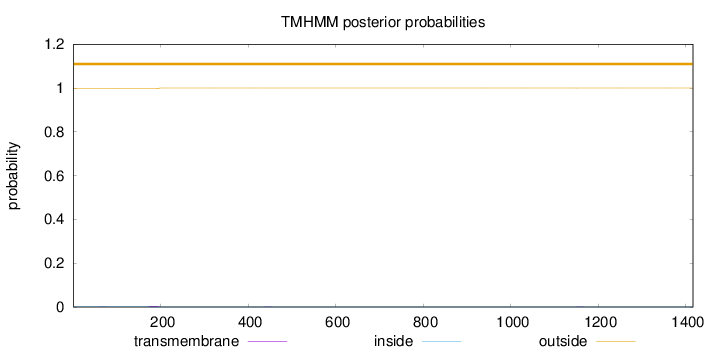
Topology
Length:
1416
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0761999999999995
Exp number, first 60 AAs:
0.0005
Total prob of N-in:
0.00333
outside
1 - 1416
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0.925507
CSRT
0
Interpretation
Uncertain
