Pre Gene Modal
BGIBMGA004054
Annotation
PREDICTED:_maspardin-like_[Amyelois_transitella]
Full name
Maspardin
Alternative Name
Spastic paraplegia 21 autosomal recessive Mast syndrome protein homolog
Location in the cell
PlasmaMembrane Reliability : 1.585
Sequence
CDS
ATGAGTTGTGCTAGCGAATTATCTCAGTCGCATGAGTATCTAAGTTTCCGAAGCAGCATTCCTTTAAGAAAAATCGTAGTAGACTCGGATAATTCCAAGGCATGGAAAATATTTGATAGTGGACCAAGGTCAGTTGCATGTCCTTTAATATGTTTACCTCCAGTTTCTGGAACAGCTGATATTTATTACAAGCAAGTTATGGGCCTAGCATCACGAGGCGTTCGAGTCATATCTGCAGAGGCTCCTCCTTATTGGAATTTAAAAGAATGGTGTGATGGTTTCAAAAGACTCATTGATATATTAGAGCTTGGTAAAGTGCACATATTCGGAGCATCTTTAGGAGGATTCATAGCCCAAAAATTTGCAGAACTGACCAGAAATTTTCCTCGTGTCGCATCACTTGTTCTGTGTAACACATTTGCGGATACGTCTGTTTTTGAATACAATGACTCAGCAGCATTATTTTGGTTACTTCCATCACTAGTTTTGAAAAGAATGTTAATGGGAAATTTTACTACTGACAAAGTCGATACACGTATGGCCGAGTCAATTGATTTTATGGTTGAAAAGCTAGAAACTCTGACGCAATCAGAATTAGCATCTAGATTGACATTGAACTGCACTCCGAATTATGTGCAACCACATTTATTAAGCGACATTCCAGTTACGATTATGGATGTTTGGGACGAAAGTGCACTTAGTTCACACGTACGTGAAGATTTGTATAAATCTTACCCTCATGCGAAGCTAGCGCATCTCAAGAGCGGTGGCAACTTTCCTTATCTTAGTAGAAGCGACGAAGTTAATTTGCATCTGTTGATACATTTAAGACAGTTTGATGGTACAGAGCTTTCGTCGTCGTATCTCAGTTTGCACAAAATGCCGCCATCCCAATCTCCCACAGACTCGGAAGAGAGCCCCATCAACTATTTTAACCAAAGTGAAAAATATGTGCAAATTTCATGA
Protein
MSCASELSQSHEYLSFRSSIPLRKIVVDSDNSKAWKIFDSGPRSVACPLICLPPVSGTADIYYKQVMGLASRGVRVISAEAPPYWNLKEWCDGFKRLIDILELGKVHIFGASLGGFIAQKFAELTRNFPRVASLVLCNTFADTSVFEYNDSAALFWLLPSLVLKRMLMGNFTTDKVDTRMAESIDFMVEKLETLTQSELASRLTLNCTPNYVQPHLLSDIPVTIMDVWDESALSSHVREDLYKSYPHAKLAHLKSGGNFPYLSRSDEVNLHLLIHLRQFDGTELSSSYLSLHKMPPSQSPTDSEESPINYFNQSEKYVQIS
Summary
Description
May play a role as a negative regulatory factor in CD4-dependent T-cell activation.
Subunit
Interacts with CD4.
Similarity
Belongs to the AB hydrolase superfamily.
Keywords
Complete proteome
Cytoplasm
Reference proteome
Phosphoprotein
Feature
chain Maspardin
Uniprot
H9J3G7
A0A2A4JA43
A0A2W1BJX3
A0A023PTS0
A0A2H1VPK8
A0A194QWL0
+ More
A0A194QQD3 A0A212EPN2 A0A154NXS6 K7IZF2 A0A2A3ERE4 A0A088AUM4 A0A0C9RN80 A0A0L7R7R7 A0A310SN11 A0A0J7KI73 A0A1W4X753 I4DKW8 A0A1Y1KNB7 A0A026VW30 A0A151X472 A0A195CNP6 F4WE58 V5I7Z6 A0A067RGW5 A0A151I5W9 A0A158NAN2 A0A151IYN0 A0A0T6AWI4 E2B4Y5 D6WQD6 A0A1Y1KJW9 A0A1B6GIW3 A0A1B6JK98 A0A1B6M629 A0A023F3Z1 A0A0K8RP44 A0A069DS48 A0A0V0G9A9 A0A224YUY1 A0A131Z339 L7LX54 T1IBN9 A0A131XEU9 A0A151JYJ7 A0A0A9YZX7 R4V3E9 A0A1E1XGI1 A0A023GBB4 A0A224XTV9 A0A1B6D4C4 A0A1B6CEJ9 E0VQU6 N6UAR4 A0A1W7RA91 A0A0P4WHT9 A7SGB1 A0A087UNC4 A0A1Z5KXD1 A0A293N5I5 T1JNR9 A0A226EVY6 F8W490 E9HMP4 F1QFZ2 H2ZSC3 H9G9D1 A0A2D0RV26 Q6PC62 K4FT91 J3SCL2 T1DKH7 A0A3Q3IVX3 A0A250YFE2 A0A0P7VTU9 L8IAZ0 Q8MJJ1 A0A098LXE7 A0A2Y9DHH4 A0A2D4FPI9 W5N9S1 A0A091KM28 A0A287D169 A0A1S3EMX4 U6CUR2 A0A2Y9KF64 M3XN72 A0A091DT71 A0A1W7RFZ6 E2QSZ2 G3SQV6 A0A2Y9HCU8 A0A2U3WKP8 A0A3Q7TPQ7 E1C0Z8 A0A0N8K1A4 F1S1X1 W5Q8G9 A0A0N8BI17 A0A341AM74
A0A194QQD3 A0A212EPN2 A0A154NXS6 K7IZF2 A0A2A3ERE4 A0A088AUM4 A0A0C9RN80 A0A0L7R7R7 A0A310SN11 A0A0J7KI73 A0A1W4X753 I4DKW8 A0A1Y1KNB7 A0A026VW30 A0A151X472 A0A195CNP6 F4WE58 V5I7Z6 A0A067RGW5 A0A151I5W9 A0A158NAN2 A0A151IYN0 A0A0T6AWI4 E2B4Y5 D6WQD6 A0A1Y1KJW9 A0A1B6GIW3 A0A1B6JK98 A0A1B6M629 A0A023F3Z1 A0A0K8RP44 A0A069DS48 A0A0V0G9A9 A0A224YUY1 A0A131Z339 L7LX54 T1IBN9 A0A131XEU9 A0A151JYJ7 A0A0A9YZX7 R4V3E9 A0A1E1XGI1 A0A023GBB4 A0A224XTV9 A0A1B6D4C4 A0A1B6CEJ9 E0VQU6 N6UAR4 A0A1W7RA91 A0A0P4WHT9 A7SGB1 A0A087UNC4 A0A1Z5KXD1 A0A293N5I5 T1JNR9 A0A226EVY6 F8W490 E9HMP4 F1QFZ2 H2ZSC3 H9G9D1 A0A2D0RV26 Q6PC62 K4FT91 J3SCL2 T1DKH7 A0A3Q3IVX3 A0A250YFE2 A0A0P7VTU9 L8IAZ0 Q8MJJ1 A0A098LXE7 A0A2Y9DHH4 A0A2D4FPI9 W5N9S1 A0A091KM28 A0A287D169 A0A1S3EMX4 U6CUR2 A0A2Y9KF64 M3XN72 A0A091DT71 A0A1W7RFZ6 E2QSZ2 G3SQV6 A0A2Y9HCU8 A0A2U3WKP8 A0A3Q7TPQ7 E1C0Z8 A0A0N8K1A4 F1S1X1 W5Q8G9 A0A0N8BI17 A0A341AM74
Pubmed
19121390
28756777
22729480
26354079
22118469
20075255
+ More
22651552 28004739 24508170 21719571 24845553 21347285 20798317 18362917 19820115 25474469 26334808 28797301 26830274 25576852 28049606 25401762 26823975 28503490 20566863 23537049 17615350 28528879 23594743 21292972 9215903 21881562 15520368 23056606 23025625 23758969 25727380 28087693 22751099 12131876 26358130 16341006 15592404 30723633 20809919
22651552 28004739 24508170 21719571 24845553 21347285 20798317 18362917 19820115 25474469 26334808 28797301 26830274 25576852 28049606 25401762 26823975 28503490 20566863 23537049 17615350 28528879 23594743 21292972 9215903 21881562 15520368 23056606 23025625 23758969 25727380 28087693 22751099 12131876 26358130 16341006 15592404 30723633 20809919
EMBL
BABH01021400
BABH01021401
BABH01021402
BABH01021403
BABH01021404
BABH01021405
+ More
BABH01021406 BABH01021407 NWSH01002390 PCG68404.1 KZ150235 PZC71973.1 JQ993446 AHX36980.1 ODYU01003670 SOQ42736.1 KQ461073 KPJ09694.1 KQ458575 KPJ05736.1 AGBW02013432 OWR43448.1 KQ434778 KZC04413.1 KZ288192 PBC34260.1 GBYB01008541 JAG78308.1 KQ414638 KOC66879.1 KQ759862 OAD62522.1 LBMM01007016 KMQ90118.1 AK401936 BAM18558.1 GEZM01081332 JAV61700.1 KK107730 EZA47967.1 KQ982557 KYQ55074.1 KQ977481 KYN02363.1 GL888102 EGI67496.1 GALX01005570 JAB62896.1 KK852646 KDR19527.1 KQ976425 KYM88100.1 ADTU01000017 KQ980763 KYN13505.1 LJIG01022678 KRT79291.1 GL445681 EFN89249.1 KQ971354 EFA06956.1 GEZM01081331 GEZM01081330 JAV61702.1 GECZ01029329 GECZ01027415 GECZ01027166 GECZ01022900 GECZ01020938 GECZ01011760 GECZ01008952 GECZ01008640 GECZ01007776 GECZ01007395 JAS40440.1 JAS42354.1 JAS42603.1 JAS46869.1 JAS48831.1 JAS58009.1 JAS60817.1 JAS61129.1 JAS61993.1 JAS62374.1 GECU01008072 JAS99634.1 GEBQ01031168 GEBQ01008595 JAT08809.1 JAT31382.1 GBBI01003038 JAC15674.1 GADI01001414 JAA72394.1 GBGD01002417 JAC86472.1 GECL01001370 JAP04754.1 GFPF01009589 MAA20735.1 GEDV01004016 JAP84541.1 GACK01008649 JAA56385.1 ACPB03006487 GEFH01003709 JAP64872.1 KQ981463 KYN41518.1 GBHO01005815 GBRD01002671 GDHC01000260 JAG37789.1 JAG63150.1 JAQ18369.1 KC632499 AGM32313.1 GFAC01000813 JAT98375.1 GBBM01003932 JAC31486.1 GFTR01004566 JAW11860.1 GEDC01016873 JAS20425.1 GEDC01025432 JAS11866.1 DS235442 EEB15752.1 APGK01042108 KB741002 KB631984 ENN75722.1 ERL87711.1 GFAH01000342 JAV48047.1 GDRN01051308 JAI66407.1 DS469651 EDO37239.1 KK120696 KFM78863.1 GFJQ02007519 JAV99450.1 GFWV01022734 MAA47461.1 AFFK01014481 LNIX01000001 OXA61773.1 CABZ01075441 CU929410 GL732688 EFX67004.1 AFYH01244460 AFYH01244461 AAWZ02026864 AY394947 BC059461 JX052336 AFK10564.1 JU175024 GBEX01001924 AFJ50550.1 JAI12636.1 GAAZ01001358 GBKC01002019 GBKD01001215 JAA96585.1 JAG44051.1 JAG46403.1 GFFW01002549 JAV42239.1 JARO02000728 KPP77588.1 JH881599 ELR53336.1 AF451182 BC118320 GBSI01001324 JAC95172.1 IACJ01087070 LAA49395.1 AHAT01024789 AHAT01024790 KK534035 KFP28869.1 AGTP01003205 AGTP01003206 HAAF01001736 CCP73562.1 AEYP01047069 AEYP01047070 AEYP01047071 KN123265 KFO25996.1 GDAY02001363 JAV50064.1 AAEX03016211 AADN05000047 JARO02001805 KPP74385.1 AEMK02000004 DQIR01095110 DQIR01246720 HDA50586.1 AMGL01101828 GDIQ01175723 JAK76002.1
BABH01021406 BABH01021407 NWSH01002390 PCG68404.1 KZ150235 PZC71973.1 JQ993446 AHX36980.1 ODYU01003670 SOQ42736.1 KQ461073 KPJ09694.1 KQ458575 KPJ05736.1 AGBW02013432 OWR43448.1 KQ434778 KZC04413.1 KZ288192 PBC34260.1 GBYB01008541 JAG78308.1 KQ414638 KOC66879.1 KQ759862 OAD62522.1 LBMM01007016 KMQ90118.1 AK401936 BAM18558.1 GEZM01081332 JAV61700.1 KK107730 EZA47967.1 KQ982557 KYQ55074.1 KQ977481 KYN02363.1 GL888102 EGI67496.1 GALX01005570 JAB62896.1 KK852646 KDR19527.1 KQ976425 KYM88100.1 ADTU01000017 KQ980763 KYN13505.1 LJIG01022678 KRT79291.1 GL445681 EFN89249.1 KQ971354 EFA06956.1 GEZM01081331 GEZM01081330 JAV61702.1 GECZ01029329 GECZ01027415 GECZ01027166 GECZ01022900 GECZ01020938 GECZ01011760 GECZ01008952 GECZ01008640 GECZ01007776 GECZ01007395 JAS40440.1 JAS42354.1 JAS42603.1 JAS46869.1 JAS48831.1 JAS58009.1 JAS60817.1 JAS61129.1 JAS61993.1 JAS62374.1 GECU01008072 JAS99634.1 GEBQ01031168 GEBQ01008595 JAT08809.1 JAT31382.1 GBBI01003038 JAC15674.1 GADI01001414 JAA72394.1 GBGD01002417 JAC86472.1 GECL01001370 JAP04754.1 GFPF01009589 MAA20735.1 GEDV01004016 JAP84541.1 GACK01008649 JAA56385.1 ACPB03006487 GEFH01003709 JAP64872.1 KQ981463 KYN41518.1 GBHO01005815 GBRD01002671 GDHC01000260 JAG37789.1 JAG63150.1 JAQ18369.1 KC632499 AGM32313.1 GFAC01000813 JAT98375.1 GBBM01003932 JAC31486.1 GFTR01004566 JAW11860.1 GEDC01016873 JAS20425.1 GEDC01025432 JAS11866.1 DS235442 EEB15752.1 APGK01042108 KB741002 KB631984 ENN75722.1 ERL87711.1 GFAH01000342 JAV48047.1 GDRN01051308 JAI66407.1 DS469651 EDO37239.1 KK120696 KFM78863.1 GFJQ02007519 JAV99450.1 GFWV01022734 MAA47461.1 AFFK01014481 LNIX01000001 OXA61773.1 CABZ01075441 CU929410 GL732688 EFX67004.1 AFYH01244460 AFYH01244461 AAWZ02026864 AY394947 BC059461 JX052336 AFK10564.1 JU175024 GBEX01001924 AFJ50550.1 JAI12636.1 GAAZ01001358 GBKC01002019 GBKD01001215 JAA96585.1 JAG44051.1 JAG46403.1 GFFW01002549 JAV42239.1 JARO02000728 KPP77588.1 JH881599 ELR53336.1 AF451182 BC118320 GBSI01001324 JAC95172.1 IACJ01087070 LAA49395.1 AHAT01024789 AHAT01024790 KK534035 KFP28869.1 AGTP01003205 AGTP01003206 HAAF01001736 CCP73562.1 AEYP01047069 AEYP01047070 AEYP01047071 KN123265 KFO25996.1 GDAY02001363 JAV50064.1 AAEX03016211 AADN05000047 JARO02001805 KPP74385.1 AEMK02000004 DQIR01095110 DQIR01246720 HDA50586.1 AMGL01101828 GDIQ01175723 JAK76002.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000076502
+ More
UP000002358 UP000242457 UP000005203 UP000053825 UP000036403 UP000192223 UP000053097 UP000075809 UP000078542 UP000007755 UP000027135 UP000078540 UP000005205 UP000078492 UP000008237 UP000007266 UP000015103 UP000078541 UP000009046 UP000019118 UP000030742 UP000001593 UP000054359 UP000198287 UP000000437 UP000000305 UP000008672 UP000001646 UP000221080 UP000261600 UP000034805 UP000192224 UP000009136 UP000248480 UP000018468 UP000005215 UP000081671 UP000248482 UP000000715 UP000028990 UP000002254 UP000007646 UP000248481 UP000245340 UP000286640 UP000000539 UP000008227 UP000002356 UP000252040
UP000002358 UP000242457 UP000005203 UP000053825 UP000036403 UP000192223 UP000053097 UP000075809 UP000078542 UP000007755 UP000027135 UP000078540 UP000005205 UP000078492 UP000008237 UP000007266 UP000015103 UP000078541 UP000009046 UP000019118 UP000030742 UP000001593 UP000054359 UP000198287 UP000000437 UP000000305 UP000008672 UP000001646 UP000221080 UP000261600 UP000034805 UP000192224 UP000009136 UP000248480 UP000018468 UP000005215 UP000081671 UP000248482 UP000000715 UP000028990 UP000002254 UP000007646 UP000248481 UP000245340 UP000286640 UP000000539 UP000008227 UP000002356 UP000252040
Pfam
PF00561 Abhydrolase_1
Interpro
Gene 3D
ProteinModelPortal
H9J3G7
A0A2A4JA43
A0A2W1BJX3
A0A023PTS0
A0A2H1VPK8
A0A194QWL0
+ More
A0A194QQD3 A0A212EPN2 A0A154NXS6 K7IZF2 A0A2A3ERE4 A0A088AUM4 A0A0C9RN80 A0A0L7R7R7 A0A310SN11 A0A0J7KI73 A0A1W4X753 I4DKW8 A0A1Y1KNB7 A0A026VW30 A0A151X472 A0A195CNP6 F4WE58 V5I7Z6 A0A067RGW5 A0A151I5W9 A0A158NAN2 A0A151IYN0 A0A0T6AWI4 E2B4Y5 D6WQD6 A0A1Y1KJW9 A0A1B6GIW3 A0A1B6JK98 A0A1B6M629 A0A023F3Z1 A0A0K8RP44 A0A069DS48 A0A0V0G9A9 A0A224YUY1 A0A131Z339 L7LX54 T1IBN9 A0A131XEU9 A0A151JYJ7 A0A0A9YZX7 R4V3E9 A0A1E1XGI1 A0A023GBB4 A0A224XTV9 A0A1B6D4C4 A0A1B6CEJ9 E0VQU6 N6UAR4 A0A1W7RA91 A0A0P4WHT9 A7SGB1 A0A087UNC4 A0A1Z5KXD1 A0A293N5I5 T1JNR9 A0A226EVY6 F8W490 E9HMP4 F1QFZ2 H2ZSC3 H9G9D1 A0A2D0RV26 Q6PC62 K4FT91 J3SCL2 T1DKH7 A0A3Q3IVX3 A0A250YFE2 A0A0P7VTU9 L8IAZ0 Q8MJJ1 A0A098LXE7 A0A2Y9DHH4 A0A2D4FPI9 W5N9S1 A0A091KM28 A0A287D169 A0A1S3EMX4 U6CUR2 A0A2Y9KF64 M3XN72 A0A091DT71 A0A1W7RFZ6 E2QSZ2 G3SQV6 A0A2Y9HCU8 A0A2U3WKP8 A0A3Q7TPQ7 E1C0Z8 A0A0N8K1A4 F1S1X1 W5Q8G9 A0A0N8BI17 A0A341AM74
A0A194QQD3 A0A212EPN2 A0A154NXS6 K7IZF2 A0A2A3ERE4 A0A088AUM4 A0A0C9RN80 A0A0L7R7R7 A0A310SN11 A0A0J7KI73 A0A1W4X753 I4DKW8 A0A1Y1KNB7 A0A026VW30 A0A151X472 A0A195CNP6 F4WE58 V5I7Z6 A0A067RGW5 A0A151I5W9 A0A158NAN2 A0A151IYN0 A0A0T6AWI4 E2B4Y5 D6WQD6 A0A1Y1KJW9 A0A1B6GIW3 A0A1B6JK98 A0A1B6M629 A0A023F3Z1 A0A0K8RP44 A0A069DS48 A0A0V0G9A9 A0A224YUY1 A0A131Z339 L7LX54 T1IBN9 A0A131XEU9 A0A151JYJ7 A0A0A9YZX7 R4V3E9 A0A1E1XGI1 A0A023GBB4 A0A224XTV9 A0A1B6D4C4 A0A1B6CEJ9 E0VQU6 N6UAR4 A0A1W7RA91 A0A0P4WHT9 A7SGB1 A0A087UNC4 A0A1Z5KXD1 A0A293N5I5 T1JNR9 A0A226EVY6 F8W490 E9HMP4 F1QFZ2 H2ZSC3 H9G9D1 A0A2D0RV26 Q6PC62 K4FT91 J3SCL2 T1DKH7 A0A3Q3IVX3 A0A250YFE2 A0A0P7VTU9 L8IAZ0 Q8MJJ1 A0A098LXE7 A0A2Y9DHH4 A0A2D4FPI9 W5N9S1 A0A091KM28 A0A287D169 A0A1S3EMX4 U6CUR2 A0A2Y9KF64 M3XN72 A0A091DT71 A0A1W7RFZ6 E2QSZ2 G3SQV6 A0A2Y9HCU8 A0A2U3WKP8 A0A3Q7TPQ7 E1C0Z8 A0A0N8K1A4 F1S1X1 W5Q8G9 A0A0N8BI17 A0A341AM74
PDB
3HZO
E-value=0.00337809,
Score=94
Ontologies
PANTHER
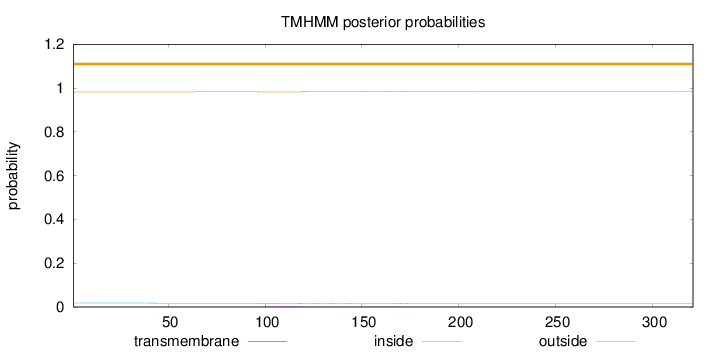
Topology
Subcellular location
Cytoplasm
Length:
321
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12469
Exp number, first 60 AAs:
0.01774
Total prob of N-in:
0.01810
outside
1 - 321
Population Genetic Test Statistics
Pi
174.762488
Theta
161.6131
Tajima's D
0.50222
CLR
0.055047
CSRT
0.518074096295185
Interpretation
Uncertain
