Pre Gene Modal
BGIBMGA004054
Annotation
PREDICTED:_tetratricopeptide_repeat_protein_19_homolog?_mitochondrial_[Bombyx_mori]
Full name
Tetratricopeptide repeat protein 19 homolog, mitochondrial
Location in the cell
Cytoplasmic Reliability : 3.104
Sequence
CDS
ATGTCAGTAAGGCTTCCTCAACAAAAGATTGGTAGTGTTCCATGTGCTGTCGGATTTTCATTGTTTACATGGCTAGGCTTCTACAAGAAAACAAGTGCTGAAGATGAACTTATACATACCATTAAACATTGTATTCTCTTCATACAGAGGTATGACTACATTAAAGCAGAACAACTATTGCATGTGGCTCTTAGACAAGCCCAACAGATGCAGCATCAGTTAGCAATAACCTATATTTATGATGTTATGGCAAACTTGGCACTTGAAAGAGAACACCTAGATAAGGCTAAAGACCTTTTTGTTGCTGTCACTCAAAGAATCATGGCTGATGGTGCAACTGAAGATGATTTTCGAGTTGTTCATATCAGTACAAAGTTAGCAAGAATAAGTCACCTGAAAAAGGATTATCAAACTGCTCAGATGGGTTATGAGTGGTGCTTAGAGAAACTAGAAAAAATTATCAAAAGCGATCCTTCAGAGGAGAAAATGAAGTTATTTGCACTAGCTGAAGATTGGTATGGACGGCTGTTCCTTGATACTAATCAATATGAACATGCCTTAAAACTTATGACCGACTCACTGGAGAAAATGCAATCCGTTTCAGATATAGAGAAAGAGCACATAGCAACTCAAATGAATGACATTGGTACAGTTTGTGATTTATTAGGTAAAACAGATGAAAGTCTACATTACTTCAAAGAAGCAATTGAAATTGGAAAAAAACTTGAAATGGAGTTAGGTACAATGTATGTGAATTTAGGAAGAGCTTACATAAAGAAAAAATTATTGTCAGAAGCAAGAAAAAATTGTGGACTGGCACTAAAAATTGGGGTCATGTCCAAAAATAAAGATGTTAAAGAGGAAGCTGAAAACTGTTTTCAAGAAATAAAAAGGTTGAATTAG
Protein
MSVRLPQQKIGSVPCAVGFSLFTWLGFYKKTSAEDELIHTIKHCILFIQRYDYIKAEQLLHVALRQAQQMQHQLAITYIYDVMANLALEREHLDKAKDLFVAVTQRIMADGATEDDFRVVHISTKLARISHLKKDYQTAQMGYEWCLEKLEKIIKSDPSEEKMKLFALAEDWYGRLFLDTNQYEHALKLMTDSLEKMQSVSDIEKEHIATQMNDIGTVCDLLGKTDESLHYFKEAIEIGKKLEMELGTMYVNLGRAYIKKKLLSEARKNCGLALKIGVMSKNKDVKEEAENCFQEIKRLN
Summary
Description
Required for mitochondrial complex III formation.
Similarity
Belongs to the TTC19 family.
Keywords
Complete proteome
Mitochondrion
Reference proteome
Repeat
TPR repeat
Transit peptide
Feature
chain Tetratricopeptide repeat protein 19 homolog, mitochondrial
Uniprot
H9J3G7
A0A2A4JAD2
A0A0L7KJV2
A0A1E1WDC5
A0A2W1BGM7
A0A3S2M6R2
+ More
S4NXZ6 A0A194QXL1 A0A194QK31 A0A2H1VQJ5 A0A212EPR8 A0A195FSW7 F4WN50 A0A195DLV4 A0A195BJA3 A0A0J7KS27 A0A158NA86 A0A026WSV9 A0A195CBG9 E2A5Q7 J3JVQ1 T1HY71 B4KE68 A0A2A3E1V5 A0A1A9WWC8 V9IEV0 A0A0A9WQV0 A0A1B6JUB4 A0A151X7P5 A0A1A9XJC3 A0A1W4XBV4 A0A1I8PUC5 A0A2R7WD61 A0A1B6F5X3 A0A1B0FF38 K7IRG7 A0A0N0U6I8 E0VVH8 A0A088AAF0 A0A1A9ZVR3 A0A1W4VXU1 J9JTK7 A0A232F8I3 A0A3B0JYR4 B4PAF6 B4LS18 A0A0A1WVY9 A0A1B0BLP1 Q29N96 B4G827 A0A2S2PGM5 A0A224XUJ9 A0A069DRH9 B3MNR1 B3NMA6 B4JAM0 A0A1Y1MET7 A0A034WPB4 A0A0J9R3V7 Q8SYD0 A0A0K8TMG2 W8BR33 B4I5Q1 A0A1B6EC26 A0A0T6AX58 B4N0Y1 B4Q9F0 A0A0K8U500 A0A1I8N193 A0A1B0GGU5 A0A139WHC1 A0A088AAF1 A0A1B6D5M8 A0A2J7QNI9 A0A2H8TW30 A0A067QS41 A0A0P4VRD4 A0A1B6LAK5 A0A1L8E0L0 A0A2S2R9X8 A0A1S3D4Q3 A0A336LP89 A0A1S4F0I6 Q17JF2 L7M1H1 A0A224Z0N5 A0A131YSF8 A0A0L7RI74 A0A182V8Q1 A0A1E1XC59 A0A182GIS5 A0A182F628 A0A084VNF1 A0A182WUB1 Q7PND9 A0A182H3P5 A0A131XE77 A0A182J605
S4NXZ6 A0A194QXL1 A0A194QK31 A0A2H1VQJ5 A0A212EPR8 A0A195FSW7 F4WN50 A0A195DLV4 A0A195BJA3 A0A0J7KS27 A0A158NA86 A0A026WSV9 A0A195CBG9 E2A5Q7 J3JVQ1 T1HY71 B4KE68 A0A2A3E1V5 A0A1A9WWC8 V9IEV0 A0A0A9WQV0 A0A1B6JUB4 A0A151X7P5 A0A1A9XJC3 A0A1W4XBV4 A0A1I8PUC5 A0A2R7WD61 A0A1B6F5X3 A0A1B0FF38 K7IRG7 A0A0N0U6I8 E0VVH8 A0A088AAF0 A0A1A9ZVR3 A0A1W4VXU1 J9JTK7 A0A232F8I3 A0A3B0JYR4 B4PAF6 B4LS18 A0A0A1WVY9 A0A1B0BLP1 Q29N96 B4G827 A0A2S2PGM5 A0A224XUJ9 A0A069DRH9 B3MNR1 B3NMA6 B4JAM0 A0A1Y1MET7 A0A034WPB4 A0A0J9R3V7 Q8SYD0 A0A0K8TMG2 W8BR33 B4I5Q1 A0A1B6EC26 A0A0T6AX58 B4N0Y1 B4Q9F0 A0A0K8U500 A0A1I8N193 A0A1B0GGU5 A0A139WHC1 A0A088AAF1 A0A1B6D5M8 A0A2J7QNI9 A0A2H8TW30 A0A067QS41 A0A0P4VRD4 A0A1B6LAK5 A0A1L8E0L0 A0A2S2R9X8 A0A1S3D4Q3 A0A336LP89 A0A1S4F0I6 Q17JF2 L7M1H1 A0A224Z0N5 A0A131YSF8 A0A0L7RI74 A0A182V8Q1 A0A1E1XC59 A0A182GIS5 A0A182F628 A0A084VNF1 A0A182WUB1 Q7PND9 A0A182H3P5 A0A131XE77 A0A182J605
Pubmed
19121390
26227816
28756777
23622113
26354079
22118469
+ More
21719571 21347285 24508170 30249741 20798317 22516182 17994087 25401762 26823975 20075255 20566863 28648823 17550304 25830018 15632085 26334808 28004739 25348373 22936249 10731132 12537572 12537569 21278747 26369729 24495485 25315136 18362917 19820115 24845553 17510324 25576852 28797301 26830274 28503490 26483478 24438588 12364791 14747013 17210077 28049606
21719571 21347285 24508170 30249741 20798317 22516182 17994087 25401762 26823975 20075255 20566863 28648823 17550304 25830018 15632085 26334808 28004739 25348373 22936249 10731132 12537572 12537569 21278747 26369729 24495485 25315136 18362917 19820115 24845553 17510324 25576852 28797301 26830274 28503490 26483478 24438588 12364791 14747013 17210077 28049606
EMBL
BABH01021400
BABH01021401
BABH01021402
BABH01021403
BABH01021404
BABH01021405
+ More
BABH01021406 BABH01021407 NWSH01002390 PCG68400.1 JTDY01009564 KOB63336.1 GDQN01006099 JAT84955.1 KZ150235 PZC71976.1 RSAL01000019 RVE52729.1 GAIX01014000 JAA78560.1 KQ461073 KPJ09695.1 KQ458575 KPJ05734.1 ODYU01003670 SOQ42732.1 AGBW02013432 OWR43451.1 KQ981305 KYN42999.1 GL888235 EGI64319.1 KQ980734 KYN13858.1 KQ976455 KYM85176.1 LBMM01003860 KMQ93044.1 ADTU01010076 KK107131 QOIP01000012 EZA58174.1 RLU15952.1 KQ978009 KYM98147.1 GL437023 EFN71254.1 BT127319 AEE62281.1 ACPB03007951 CH933807 EDW11813.1 KZ288444 PBC25677.1 JR042580 AEY59638.1 GBHO01036384 GBHO01036372 GBHO01000865 GBHO01000862 GBRD01004314 GDHC01006760 JAG07220.1 JAG07232.1 JAG42739.1 JAG42742.1 JAG61507.1 JAQ11869.1 GECU01035006 GECU01032592 GECU01029209 GECU01029008 GECU01011403 GECU01005327 GECU01004909 GECU01004658 JAS72700.1 JAS75114.1 JAS78497.1 JAS78698.1 JAS96303.1 JAT02380.1 JAT02798.1 JAT03049.1 KQ982446 KYQ56344.1 KK854483 PTY16255.1 GECZ01024110 JAS45659.1 CCAG010009466 KQ435727 KOX77971.1 DS235812 EEB17384.1 ABLF02035184 NNAY01000719 OXU26800.1 OUUW01000010 SPP86213.1 CM000158 EDW90364.1 CH940649 EDW63694.1 GBXI01011612 JAD02680.1 JXJN01016521 CH379060 EAL33446.2 CH479180 EDW28507.1 GGMR01015970 MBY28589.1 GFTR01004643 JAW11783.1 GBGD01002374 JAC86515.1 CH902620 EDV31148.1 CH954179 EDV54706.2 CH916368 EDW02806.1 GEZM01033223 JAV84339.1 GAKP01001531 JAC57421.1 CM002910 KMY90927.1 AE014134 AY071630 GDAI01002267 JAI15336.1 GAMC01005043 GAMC01005041 JAC01513.1 CH480822 EDW55707.1 GEDC01001874 JAS35424.1 LJIG01022601 KRT79706.1 CH963920 EDW78143.1 CM000361 EDX05457.1 GDHF01030864 JAI21450.1 AJWK01000755 KQ971343 KYB27187.1 GEDC01016311 JAS20987.1 NEVH01013194 PNF30154.1 GFXV01006652 MBW18457.1 KK853366 KDR08095.1 GDRN01104120 GDRN01104119 JAI57967.1 GEBQ01019326 JAT20651.1 GFDF01002049 JAV12035.1 GGMS01017570 MBY86773.1 UFQT01000037 SSX18369.1 CH477232 EAT46888.1 GACK01007129 JAA57905.1 GFPF01012280 MAA23426.1 GEDV01007716 JAP80841.1 KQ414585 KOC70529.1 GFAC01002522 JAT96666.1 JXUM01012160 KQ560347 KXJ82916.1 ATLV01014738 KE524984 KFB39495.1 AAAB01008964 EAA12340.5 JXUM01024539 KQ560693 KXJ81251.1 GEFH01003919 JAP64662.1
BABH01021406 BABH01021407 NWSH01002390 PCG68400.1 JTDY01009564 KOB63336.1 GDQN01006099 JAT84955.1 KZ150235 PZC71976.1 RSAL01000019 RVE52729.1 GAIX01014000 JAA78560.1 KQ461073 KPJ09695.1 KQ458575 KPJ05734.1 ODYU01003670 SOQ42732.1 AGBW02013432 OWR43451.1 KQ981305 KYN42999.1 GL888235 EGI64319.1 KQ980734 KYN13858.1 KQ976455 KYM85176.1 LBMM01003860 KMQ93044.1 ADTU01010076 KK107131 QOIP01000012 EZA58174.1 RLU15952.1 KQ978009 KYM98147.1 GL437023 EFN71254.1 BT127319 AEE62281.1 ACPB03007951 CH933807 EDW11813.1 KZ288444 PBC25677.1 JR042580 AEY59638.1 GBHO01036384 GBHO01036372 GBHO01000865 GBHO01000862 GBRD01004314 GDHC01006760 JAG07220.1 JAG07232.1 JAG42739.1 JAG42742.1 JAG61507.1 JAQ11869.1 GECU01035006 GECU01032592 GECU01029209 GECU01029008 GECU01011403 GECU01005327 GECU01004909 GECU01004658 JAS72700.1 JAS75114.1 JAS78497.1 JAS78698.1 JAS96303.1 JAT02380.1 JAT02798.1 JAT03049.1 KQ982446 KYQ56344.1 KK854483 PTY16255.1 GECZ01024110 JAS45659.1 CCAG010009466 KQ435727 KOX77971.1 DS235812 EEB17384.1 ABLF02035184 NNAY01000719 OXU26800.1 OUUW01000010 SPP86213.1 CM000158 EDW90364.1 CH940649 EDW63694.1 GBXI01011612 JAD02680.1 JXJN01016521 CH379060 EAL33446.2 CH479180 EDW28507.1 GGMR01015970 MBY28589.1 GFTR01004643 JAW11783.1 GBGD01002374 JAC86515.1 CH902620 EDV31148.1 CH954179 EDV54706.2 CH916368 EDW02806.1 GEZM01033223 JAV84339.1 GAKP01001531 JAC57421.1 CM002910 KMY90927.1 AE014134 AY071630 GDAI01002267 JAI15336.1 GAMC01005043 GAMC01005041 JAC01513.1 CH480822 EDW55707.1 GEDC01001874 JAS35424.1 LJIG01022601 KRT79706.1 CH963920 EDW78143.1 CM000361 EDX05457.1 GDHF01030864 JAI21450.1 AJWK01000755 KQ971343 KYB27187.1 GEDC01016311 JAS20987.1 NEVH01013194 PNF30154.1 GFXV01006652 MBW18457.1 KK853366 KDR08095.1 GDRN01104120 GDRN01104119 JAI57967.1 GEBQ01019326 JAT20651.1 GFDF01002049 JAV12035.1 GGMS01017570 MBY86773.1 UFQT01000037 SSX18369.1 CH477232 EAT46888.1 GACK01007129 JAA57905.1 GFPF01012280 MAA23426.1 GEDV01007716 JAP80841.1 KQ414585 KOC70529.1 GFAC01002522 JAT96666.1 JXUM01012160 KQ560347 KXJ82916.1 ATLV01014738 KE524984 KFB39495.1 AAAB01008964 EAA12340.5 JXUM01024539 KQ560693 KXJ81251.1 GEFH01003919 JAP64662.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000053240
UP000053268
+ More
UP000007151 UP000078541 UP000007755 UP000078492 UP000078540 UP000036403 UP000005205 UP000053097 UP000279307 UP000078542 UP000000311 UP000015103 UP000009192 UP000242457 UP000091820 UP000075809 UP000092443 UP000192223 UP000095300 UP000092444 UP000002358 UP000053105 UP000009046 UP000005203 UP000092445 UP000192221 UP000007819 UP000215335 UP000268350 UP000002282 UP000008792 UP000092460 UP000001819 UP000008744 UP000007801 UP000008711 UP000001070 UP000000803 UP000001292 UP000007798 UP000000304 UP000095301 UP000092461 UP000007266 UP000235965 UP000027135 UP000079169 UP000008820 UP000053825 UP000075903 UP000069940 UP000249989 UP000069272 UP000030765 UP000076407 UP000007062 UP000075880
UP000007151 UP000078541 UP000007755 UP000078492 UP000078540 UP000036403 UP000005205 UP000053097 UP000279307 UP000078542 UP000000311 UP000015103 UP000009192 UP000242457 UP000091820 UP000075809 UP000092443 UP000192223 UP000095300 UP000092444 UP000002358 UP000053105 UP000009046 UP000005203 UP000092445 UP000192221 UP000007819 UP000215335 UP000268350 UP000002282 UP000008792 UP000092460 UP000001819 UP000008744 UP000007801 UP000008711 UP000001070 UP000000803 UP000001292 UP000007798 UP000000304 UP000095301 UP000092461 UP000007266 UP000235965 UP000027135 UP000079169 UP000008820 UP000053825 UP000075903 UP000069940 UP000249989 UP000069272 UP000030765 UP000076407 UP000007062 UP000075880
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9J3G7
A0A2A4JAD2
A0A0L7KJV2
A0A1E1WDC5
A0A2W1BGM7
A0A3S2M6R2
+ More
S4NXZ6 A0A194QXL1 A0A194QK31 A0A2H1VQJ5 A0A212EPR8 A0A195FSW7 F4WN50 A0A195DLV4 A0A195BJA3 A0A0J7KS27 A0A158NA86 A0A026WSV9 A0A195CBG9 E2A5Q7 J3JVQ1 T1HY71 B4KE68 A0A2A3E1V5 A0A1A9WWC8 V9IEV0 A0A0A9WQV0 A0A1B6JUB4 A0A151X7P5 A0A1A9XJC3 A0A1W4XBV4 A0A1I8PUC5 A0A2R7WD61 A0A1B6F5X3 A0A1B0FF38 K7IRG7 A0A0N0U6I8 E0VVH8 A0A088AAF0 A0A1A9ZVR3 A0A1W4VXU1 J9JTK7 A0A232F8I3 A0A3B0JYR4 B4PAF6 B4LS18 A0A0A1WVY9 A0A1B0BLP1 Q29N96 B4G827 A0A2S2PGM5 A0A224XUJ9 A0A069DRH9 B3MNR1 B3NMA6 B4JAM0 A0A1Y1MET7 A0A034WPB4 A0A0J9R3V7 Q8SYD0 A0A0K8TMG2 W8BR33 B4I5Q1 A0A1B6EC26 A0A0T6AX58 B4N0Y1 B4Q9F0 A0A0K8U500 A0A1I8N193 A0A1B0GGU5 A0A139WHC1 A0A088AAF1 A0A1B6D5M8 A0A2J7QNI9 A0A2H8TW30 A0A067QS41 A0A0P4VRD4 A0A1B6LAK5 A0A1L8E0L0 A0A2S2R9X8 A0A1S3D4Q3 A0A336LP89 A0A1S4F0I6 Q17JF2 L7M1H1 A0A224Z0N5 A0A131YSF8 A0A0L7RI74 A0A182V8Q1 A0A1E1XC59 A0A182GIS5 A0A182F628 A0A084VNF1 A0A182WUB1 Q7PND9 A0A182H3P5 A0A131XE77 A0A182J605
S4NXZ6 A0A194QXL1 A0A194QK31 A0A2H1VQJ5 A0A212EPR8 A0A195FSW7 F4WN50 A0A195DLV4 A0A195BJA3 A0A0J7KS27 A0A158NA86 A0A026WSV9 A0A195CBG9 E2A5Q7 J3JVQ1 T1HY71 B4KE68 A0A2A3E1V5 A0A1A9WWC8 V9IEV0 A0A0A9WQV0 A0A1B6JUB4 A0A151X7P5 A0A1A9XJC3 A0A1W4XBV4 A0A1I8PUC5 A0A2R7WD61 A0A1B6F5X3 A0A1B0FF38 K7IRG7 A0A0N0U6I8 E0VVH8 A0A088AAF0 A0A1A9ZVR3 A0A1W4VXU1 J9JTK7 A0A232F8I3 A0A3B0JYR4 B4PAF6 B4LS18 A0A0A1WVY9 A0A1B0BLP1 Q29N96 B4G827 A0A2S2PGM5 A0A224XUJ9 A0A069DRH9 B3MNR1 B3NMA6 B4JAM0 A0A1Y1MET7 A0A034WPB4 A0A0J9R3V7 Q8SYD0 A0A0K8TMG2 W8BR33 B4I5Q1 A0A1B6EC26 A0A0T6AX58 B4N0Y1 B4Q9F0 A0A0K8U500 A0A1I8N193 A0A1B0GGU5 A0A139WHC1 A0A088AAF1 A0A1B6D5M8 A0A2J7QNI9 A0A2H8TW30 A0A067QS41 A0A0P4VRD4 A0A1B6LAK5 A0A1L8E0L0 A0A2S2R9X8 A0A1S3D4Q3 A0A336LP89 A0A1S4F0I6 Q17JF2 L7M1H1 A0A224Z0N5 A0A131YSF8 A0A0L7RI74 A0A182V8Q1 A0A1E1XC59 A0A182GIS5 A0A182F628 A0A084VNF1 A0A182WUB1 Q7PND9 A0A182H3P5 A0A131XE77 A0A182J605
PDB
5O01
E-value=0.00328251,
Score=94
Ontologies
GO
Topology
Subcellular location
Mitochondrion
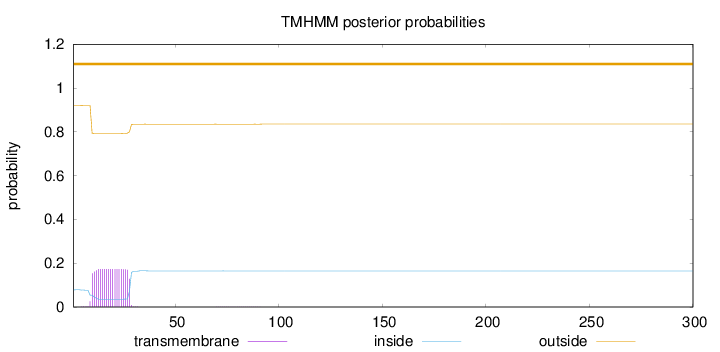
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.26366
Exp number, first 60 AAs:
3.25104
Total prob of N-in:
0.07844
outside
1 - 300
Population Genetic Test Statistics
Pi
145.131813
Theta
187.682209
Tajima's D
-0.713439
CLR
557.242068
CSRT
0.196190190490475
Interpretation
Uncertain
