Gene
KWMTBOMO11511 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004059
Annotation
peptidylprolyl_isomerase_B_precursor_[Bombyx_mori]
Full name
Peptidyl-prolyl cis-trans isomerase
+ More
Peptidylprolyl isomerase
Peptidylprolyl isomerase
Location in the cell
Cytoplasmic Reliability : 3.087
Sequence
CDS
ATGGGTACCCTTACAATGGCTTTAGGAATTTTGTTGTTCATCGCTAGTGCCAAATCTGATGAAATTCCCAAAGGACCTAAAGTTACTCATAAGGTGAGCTTTGACATGAAGATTGGTGATGACAATATTGGTACTATTGTGATTGGATTATTTGGAAAGACTGTACCTAAGACAACTGAGAACTTCTTTCAACTAGCTCAGAAACCTGAGGGGGAGGGGTACAAAGGGAGCAAGTTCCACAGAGTAATTAAAAATTTCATGATCCAAGGTGGTGATTTTACCAAGGGTGATGGAACTGGAGGGCGCAGTATATATGGTGAACGTTTTGAAGATGAAAACTTCAAGCTGAAGCACTATGGTGCTGGTTGGTTATCTATGGCTAATGCAGGCAAAGACACAAATGGATCTCAATTTTTCATCACAACTGTTAAGACACCCTGGTTAGATGGCAGACATGTTGTTTTCGGTAAAGTTTTAGAAGGAATGGATGTTGTACAGAAAATTGAGATGACTGTTACGGGTGCGAATGATCGCCCAGTCAAAGATGTTGTTATATCTGACACGAAAACTGAAGTTGTAGCTGAACCTTTCAGTGTCACAAAAGAGAGTGCTCACTAA
Protein
MGTLTMALGILLFIASAKSDEIPKGPKVTHKVSFDMKIGDDNIGTIVIGLFGKTVPKTTENFFQLAQKPEGEGYKGSKFHRVIKNFMIQGGDFTKGDGTGGRSIYGERFEDENFKLKHYGAGWLSMANAGKDTNGSQFFITTVKTPWLDGRHVVFGKVLEGMDVVQKIEMTVTGANDRPVKDVVISDTKTEVVAEPFSVTKESAH
Summary
Description
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides.
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Similarity
Belongs to the cyclophilin-type PPIase family.
Feature
chain Peptidyl-prolyl cis-trans isomerase
Uniprot
Q1HPL6
B2LU30
H9J3H2
A0A2H1VPI2
A0A2W1BDP4
A0A2A4J8W7
+ More
A0A194R1H0 I4DIV0 A0A194QJI4 A0A212EPM9 I4DMJ8 S4NSY7 A0A0L7KJQ9 D6WP73 A0A1W4WZJ5 E2B9N3 A0A0U4CK71 V5IA37 A0A026VU23 A0A085NKC6 A0A085M534 E9IVJ0 A0A348G6B4 A0A151IXN8 R4WNH2 A0A195B3U7 A0A0U2KDH9 T1PFJ9 A0A146LEZ1 A0A1I8PL32 A0A0M9A7P1 E0VQ50 A0A1W4VQI2 A0A158P494 A0A2J7R9C7 F6NLI6 Q6PBW4 A0A1D1Y996 A0A077Z7S3 A0A232FK40 A0A2P0XIY7 B4LMM9 A0A3P9JGA9 H2L8F1 A0A0J7KM63 U4U7D9 A0A3B3C2S8 A0A067QW87 B4KTE5 Q28G13 F7B346 E2ADG5 A0A0A9VZ13 A8XWF8 A0A0A9VUP2 A0A3N0Z8L6 A0A2G5VST2 A0A2H2JGU3 A0A1A7XIV7 A0A0M5J582 F4WIU3 B4J4H9 A0A3B4DBR6 Q2I190 A0A1A8RNU8 A0A1A8JZQ9 A0A1A8NKH1 A0A1A8CBK8 A0A1A8H677 A0A1A7ZX97 E4YTB2 B4MQG3 A0A3Q2XRH9 E9FQX5 A0A0L0BPH9 A0A1L8EBX2 B3MEC1 A0A195CTY2 A0A2I4CR27 A0A165A8N4 A0A0N5AZ53 A0A0T6B2I3 A0A3B4WEA3 A0A3B4VP14 A0A3Q3JPU1 A0A0P5HJR9 A0A3P9LJK0 A0A3Q0T794 A0A0P5HS88 A0A0F8D2N6 A0A0U2T4N8 A8C9X3 A0A3Q3T1C1 A0A0P5GG94 A0A0N8AJ41 A0A0P5EL65 A0A1D2N880 I3K4T4 A0A2Z4EE38 E4XG60
A0A194R1H0 I4DIV0 A0A194QJI4 A0A212EPM9 I4DMJ8 S4NSY7 A0A0L7KJQ9 D6WP73 A0A1W4WZJ5 E2B9N3 A0A0U4CK71 V5IA37 A0A026VU23 A0A085NKC6 A0A085M534 E9IVJ0 A0A348G6B4 A0A151IXN8 R4WNH2 A0A195B3U7 A0A0U2KDH9 T1PFJ9 A0A146LEZ1 A0A1I8PL32 A0A0M9A7P1 E0VQ50 A0A1W4VQI2 A0A158P494 A0A2J7R9C7 F6NLI6 Q6PBW4 A0A1D1Y996 A0A077Z7S3 A0A232FK40 A0A2P0XIY7 B4LMM9 A0A3P9JGA9 H2L8F1 A0A0J7KM63 U4U7D9 A0A3B3C2S8 A0A067QW87 B4KTE5 Q28G13 F7B346 E2ADG5 A0A0A9VZ13 A8XWF8 A0A0A9VUP2 A0A3N0Z8L6 A0A2G5VST2 A0A2H2JGU3 A0A1A7XIV7 A0A0M5J582 F4WIU3 B4J4H9 A0A3B4DBR6 Q2I190 A0A1A8RNU8 A0A1A8JZQ9 A0A1A8NKH1 A0A1A8CBK8 A0A1A8H677 A0A1A7ZX97 E4YTB2 B4MQG3 A0A3Q2XRH9 E9FQX5 A0A0L0BPH9 A0A1L8EBX2 B3MEC1 A0A195CTY2 A0A2I4CR27 A0A165A8N4 A0A0N5AZ53 A0A0T6B2I3 A0A3B4WEA3 A0A3B4VP14 A0A3Q3JPU1 A0A0P5HJR9 A0A3P9LJK0 A0A3Q0T794 A0A0P5HS88 A0A0F8D2N6 A0A0U2T4N8 A8C9X3 A0A3Q3T1C1 A0A0P5GG94 A0A0N8AJ41 A0A0P5EL65 A0A1D2N880 I3K4T4 A0A2Z4EE38 E4XG60
EC Number
5.2.1.8
Pubmed
19121390
28756777
26354079
22651552
22118469
23622113
+ More
26227816 18362917 19820115 20798317 26694822 24508170 30249741 24929829 21282665 23691247 26621068 25315136 26823975 20566863 21347285 23594743 12477932 28648823 17994087 17554307 23537049 29451363 24845553 20431018 25401762 14624247 21719571 18001845 21097902 21292972 26108605 25835551 17760985 27289101 25186727
26227816 18362917 19820115 20798317 26694822 24508170 30249741 24929829 21282665 23691247 26621068 25315136 26823975 20566863 21347285 23594743 12477932 28648823 17994087 17554307 23537049 29451363 24845553 20431018 25401762 14624247 21719571 18001845 21097902 21292972 26108605 25835551 17760985 27289101 25186727
EMBL
DQ443386
ABF51475.1
EU583493
ACB72457.1
BABH01021399
BABH01021400
+ More
ODYU01003670 SOQ42733.1 KZ150235 PZC71975.1 NWSH01002390 PCG68401.1 KQ461073 KPJ09696.1 AK401218 BAM17840.1 KQ458575 KPJ05733.1 AGBW02013432 OWR43452.1 AK402516 BAM19138.1 GAIX01012381 JAA80179.1 JTDY01009564 KOB63335.1 KQ971352 EFA07271.1 GL446584 EFN87567.1 KU218652 ALX00032.1 GALX01001865 JAB66601.1 KK107928 QOIP01000001 EZA47135.1 RLU26552.1 KL367491 KFD69922.1 KL363228 KN538417 KFD52330.1 KHJ41401.1 GL766258 EFZ15398.1 FX986030 BBF97987.1 KQ980796 KYN13004.1 AK417181 BAN20396.1 KQ976618 KYM79163.1 KT754848 ALS04682.1 KA647511 AFP62140.1 GDHC01012470 JAQ06159.1 KQ435732 KOX77549.1 DS235389 EEB15506.1 ADTU01008749 NEVH01006598 PNF37436.1 AL929134 BC059560 BC071458 BC153469 BC154213 AAH59560.1 AAH71458.1 AAI53470.1 AAI54214.1 GDJX01016740 JAT51196.1 HG806021 CDW56256.1 NNAY01000089 OXU31106.1 KY661312 AVA17399.1 CH940648 EDW60016.1 LBMM01005605 KMQ91382.1 KB632046 ERL88263.1 KK852942 KDR13508.1 CH933808 EDW08506.1 CR761630 CAJ82504.2 AAMC01085641 GL438750 EFN68527.1 GBHO01044181 GBRD01009958 JAF99422.1 JAG55866.1 HE601474 CAP36977.1 GBHO01044180 JAF99423.1 RJVU01006614 ROL54671.1 PDUG01000001 PIC54646.1 HADW01016592 HADX01005380 SBP17992.1 CP012524 ALC42381.1 GL888177 EGI65889.1 CH916367 EDW01661.1 DQ353764 EF424275 ABC75555.1 ABO15710.1 HAEH01007995 HAEI01009294 SBS07367.1 HAED01015143 HAEE01005446 SBR25466.1 HAEF01015094 HAEG01003474 SBR69302.1 HADZ01012194 HAEA01006136 SBP76135.1 HAEB01007898 HAEC01010611 SBQ78827.1 HADY01009027 HAEJ01004464 SBP47512.1 FN655300 CBY38701.1 CH963849 EDW74352.1 GL732523 EFX90061.1 JRES01001578 KNC21868.1 GFDG01002562 JAV16237.1 CH902619 EDV36527.1 KQ977279 KYN04146.1 LRGB01000642 KZS17305.1 LJIG01016196 KRT81326.1 GDIQ01232116 JAK19609.1 GDIQ01223665 JAK28060.1 KQ040883 KKF33456.1 KT754615 ALS04449.1 EU032351 ABV44718.1 GDIQ01242815 JAK08910.1 GDIQ01270731 JAJ80993.1 GDIQ01270335 JAJ81389.1 LJIJ01000150 ODN01473.1 AERX01017850 AERX01017851 MG722837 AWV50529.1 FN653047 CBY09658.1
ODYU01003670 SOQ42733.1 KZ150235 PZC71975.1 NWSH01002390 PCG68401.1 KQ461073 KPJ09696.1 AK401218 BAM17840.1 KQ458575 KPJ05733.1 AGBW02013432 OWR43452.1 AK402516 BAM19138.1 GAIX01012381 JAA80179.1 JTDY01009564 KOB63335.1 KQ971352 EFA07271.1 GL446584 EFN87567.1 KU218652 ALX00032.1 GALX01001865 JAB66601.1 KK107928 QOIP01000001 EZA47135.1 RLU26552.1 KL367491 KFD69922.1 KL363228 KN538417 KFD52330.1 KHJ41401.1 GL766258 EFZ15398.1 FX986030 BBF97987.1 KQ980796 KYN13004.1 AK417181 BAN20396.1 KQ976618 KYM79163.1 KT754848 ALS04682.1 KA647511 AFP62140.1 GDHC01012470 JAQ06159.1 KQ435732 KOX77549.1 DS235389 EEB15506.1 ADTU01008749 NEVH01006598 PNF37436.1 AL929134 BC059560 BC071458 BC153469 BC154213 AAH59560.1 AAH71458.1 AAI53470.1 AAI54214.1 GDJX01016740 JAT51196.1 HG806021 CDW56256.1 NNAY01000089 OXU31106.1 KY661312 AVA17399.1 CH940648 EDW60016.1 LBMM01005605 KMQ91382.1 KB632046 ERL88263.1 KK852942 KDR13508.1 CH933808 EDW08506.1 CR761630 CAJ82504.2 AAMC01085641 GL438750 EFN68527.1 GBHO01044181 GBRD01009958 JAF99422.1 JAG55866.1 HE601474 CAP36977.1 GBHO01044180 JAF99423.1 RJVU01006614 ROL54671.1 PDUG01000001 PIC54646.1 HADW01016592 HADX01005380 SBP17992.1 CP012524 ALC42381.1 GL888177 EGI65889.1 CH916367 EDW01661.1 DQ353764 EF424275 ABC75555.1 ABO15710.1 HAEH01007995 HAEI01009294 SBS07367.1 HAED01015143 HAEE01005446 SBR25466.1 HAEF01015094 HAEG01003474 SBR69302.1 HADZ01012194 HAEA01006136 SBP76135.1 HAEB01007898 HAEC01010611 SBQ78827.1 HADY01009027 HAEJ01004464 SBP47512.1 FN655300 CBY38701.1 CH963849 EDW74352.1 GL732523 EFX90061.1 JRES01001578 KNC21868.1 GFDG01002562 JAV16237.1 CH902619 EDV36527.1 KQ977279 KYN04146.1 LRGB01000642 KZS17305.1 LJIG01016196 KRT81326.1 GDIQ01232116 JAK19609.1 GDIQ01223665 JAK28060.1 KQ040883 KKF33456.1 KT754615 ALS04449.1 EU032351 ABV44718.1 GDIQ01242815 JAK08910.1 GDIQ01270731 JAJ80993.1 GDIQ01270335 JAJ81389.1 LJIJ01000150 ODN01473.1 AERX01017850 AERX01017851 MG722837 AWV50529.1 FN653047 CBY09658.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000007266 UP000192223 UP000008237 UP000053097 UP000279307 UP000078492 UP000078540 UP000095301 UP000095300 UP000053105 UP000009046 UP000192221 UP000005205 UP000235965 UP000000437 UP000030665 UP000215335 UP000008792 UP000265200 UP000001038 UP000036403 UP000030742 UP000261560 UP000027135 UP000009192 UP000008143 UP000000311 UP000008549 UP000230233 UP000005237 UP000092553 UP000007755 UP000001070 UP000261440 UP000221080 UP000007798 UP000264820 UP000000305 UP000037069 UP000007801 UP000078542 UP000192220 UP000076858 UP000046393 UP000261360 UP000261420 UP000261600 UP000265180 UP000261340 UP000261640 UP000094527 UP000005207
UP000007266 UP000192223 UP000008237 UP000053097 UP000279307 UP000078492 UP000078540 UP000095301 UP000095300 UP000053105 UP000009046 UP000192221 UP000005205 UP000235965 UP000000437 UP000030665 UP000215335 UP000008792 UP000265200 UP000001038 UP000036403 UP000030742 UP000261560 UP000027135 UP000009192 UP000008143 UP000000311 UP000008549 UP000230233 UP000005237 UP000092553 UP000007755 UP000001070 UP000261440 UP000221080 UP000007798 UP000264820 UP000000305 UP000037069 UP000007801 UP000078542 UP000192220 UP000076858 UP000046393 UP000261360 UP000261420 UP000261600 UP000265180 UP000261340 UP000261640 UP000094527 UP000005207
Interpro
Gene 3D
ProteinModelPortal
Q1HPL6
B2LU30
H9J3H2
A0A2H1VPI2
A0A2W1BDP4
A0A2A4J8W7
+ More
A0A194R1H0 I4DIV0 A0A194QJI4 A0A212EPM9 I4DMJ8 S4NSY7 A0A0L7KJQ9 D6WP73 A0A1W4WZJ5 E2B9N3 A0A0U4CK71 V5IA37 A0A026VU23 A0A085NKC6 A0A085M534 E9IVJ0 A0A348G6B4 A0A151IXN8 R4WNH2 A0A195B3U7 A0A0U2KDH9 T1PFJ9 A0A146LEZ1 A0A1I8PL32 A0A0M9A7P1 E0VQ50 A0A1W4VQI2 A0A158P494 A0A2J7R9C7 F6NLI6 Q6PBW4 A0A1D1Y996 A0A077Z7S3 A0A232FK40 A0A2P0XIY7 B4LMM9 A0A3P9JGA9 H2L8F1 A0A0J7KM63 U4U7D9 A0A3B3C2S8 A0A067QW87 B4KTE5 Q28G13 F7B346 E2ADG5 A0A0A9VZ13 A8XWF8 A0A0A9VUP2 A0A3N0Z8L6 A0A2G5VST2 A0A2H2JGU3 A0A1A7XIV7 A0A0M5J582 F4WIU3 B4J4H9 A0A3B4DBR6 Q2I190 A0A1A8RNU8 A0A1A8JZQ9 A0A1A8NKH1 A0A1A8CBK8 A0A1A8H677 A0A1A7ZX97 E4YTB2 B4MQG3 A0A3Q2XRH9 E9FQX5 A0A0L0BPH9 A0A1L8EBX2 B3MEC1 A0A195CTY2 A0A2I4CR27 A0A165A8N4 A0A0N5AZ53 A0A0T6B2I3 A0A3B4WEA3 A0A3B4VP14 A0A3Q3JPU1 A0A0P5HJR9 A0A3P9LJK0 A0A3Q0T794 A0A0P5HS88 A0A0F8D2N6 A0A0U2T4N8 A8C9X3 A0A3Q3T1C1 A0A0P5GG94 A0A0N8AJ41 A0A0P5EL65 A0A1D2N880 I3K4T4 A0A2Z4EE38 E4XG60
A0A194R1H0 I4DIV0 A0A194QJI4 A0A212EPM9 I4DMJ8 S4NSY7 A0A0L7KJQ9 D6WP73 A0A1W4WZJ5 E2B9N3 A0A0U4CK71 V5IA37 A0A026VU23 A0A085NKC6 A0A085M534 E9IVJ0 A0A348G6B4 A0A151IXN8 R4WNH2 A0A195B3U7 A0A0U2KDH9 T1PFJ9 A0A146LEZ1 A0A1I8PL32 A0A0M9A7P1 E0VQ50 A0A1W4VQI2 A0A158P494 A0A2J7R9C7 F6NLI6 Q6PBW4 A0A1D1Y996 A0A077Z7S3 A0A232FK40 A0A2P0XIY7 B4LMM9 A0A3P9JGA9 H2L8F1 A0A0J7KM63 U4U7D9 A0A3B3C2S8 A0A067QW87 B4KTE5 Q28G13 F7B346 E2ADG5 A0A0A9VZ13 A8XWF8 A0A0A9VUP2 A0A3N0Z8L6 A0A2G5VST2 A0A2H2JGU3 A0A1A7XIV7 A0A0M5J582 F4WIU3 B4J4H9 A0A3B4DBR6 Q2I190 A0A1A8RNU8 A0A1A8JZQ9 A0A1A8NKH1 A0A1A8CBK8 A0A1A8H677 A0A1A7ZX97 E4YTB2 B4MQG3 A0A3Q2XRH9 E9FQX5 A0A0L0BPH9 A0A1L8EBX2 B3MEC1 A0A195CTY2 A0A2I4CR27 A0A165A8N4 A0A0N5AZ53 A0A0T6B2I3 A0A3B4WEA3 A0A3B4VP14 A0A3Q3JPU1 A0A0P5HJR9 A0A3P9LJK0 A0A3Q0T794 A0A0P5HS88 A0A0F8D2N6 A0A0U2T4N8 A8C9X3 A0A3Q3T1C1 A0A0P5GG94 A0A0N8AJ41 A0A0P5EL65 A0A1D2N880 I3K4T4 A0A2Z4EE38 E4XG60
PDB
1H0P
E-value=1.5776e-64,
Score=621
Ontologies
GO
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.994492
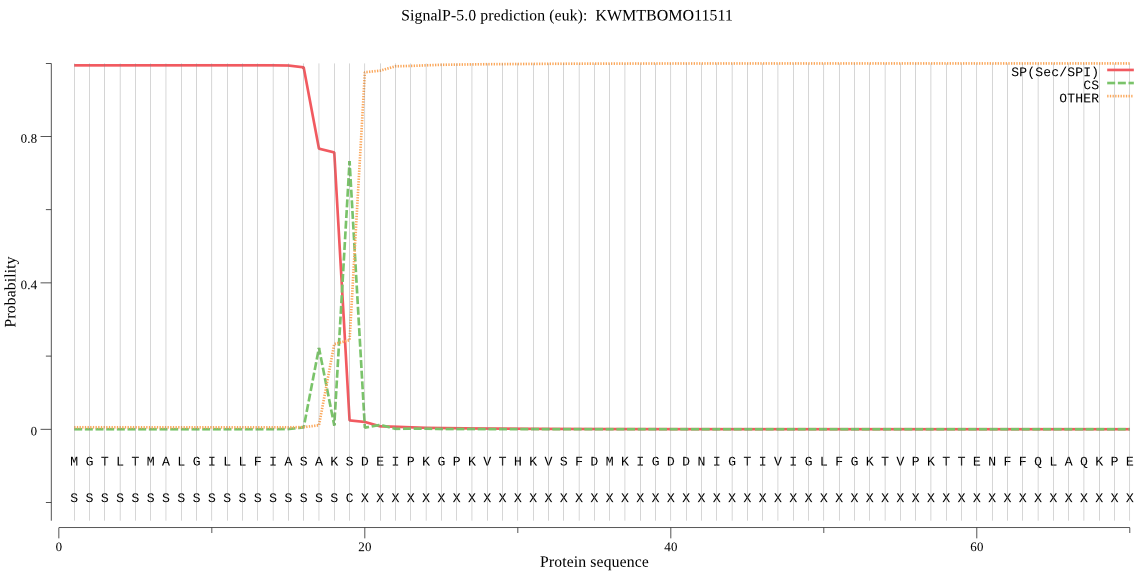
Length:
205
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00761999999999999
Exp number, first 60 AAs:
0.00741
Total prob of N-in:
0.02659
outside
1 - 205

Population Genetic Test Statistics
Pi
131.821075
Theta
155.48049
Tajima's D
-0.498013
CLR
0.807453
CSRT
0.240137993100345
Interpretation
Uncertain
