Gene
KWMTBOMO11508 Validated by peptides from experiments
Annotation
PREDICTED:_pentatricopeptide_repeat-containing_protein_2?_mitochondrial-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.738 Nuclear Reliability : 1.075
Sequence
CDS
ATGTCTCTGTTTCTAAATTATTTTTTGAAGACTTCTTTCAGCTGTAATCAACGATTGTTTTTACCCTGCAGGCAATTACATTCTACACAAGTGAGGCTGCTGTATGCCCCTGCAACAATTGGAATAGATGGGTATTTACAGGCCAGGAAAAATGTCAAAGAACAATTTGTCAATTTCTCTGAGAAATTTAAAACTAAAATGAATGATTTTGTCTCCGATTCCAAAAACATGATATTCACTGAAGATTTGAAGAACATGGTTCATTTGGCAGAACCATCAGACTTGCAACTTGTACTAAATATGATTAAAAAATTCAACACACAAAGTACAGAGTTCAGATTTGGTTCATTCGTGTTTGGCCCAGTTGTAATGAGGATGTTTCATTTCTTAGATGCTCCTAAAGAAGCACTGCAGTGTTTTGAAGAACCAGCAAATGATGGTTTTTTTGATCAACAAGTGTCATACCAGATATTACTTGATTTACTGTATTGTCATGAAATGTTTGATGAGATGTACAGGGTCTTTGAGAAAGTACAAAAGAAACAAATAAATATGACAAAGTTTCCTAAATACTGTGTGGTGTTGATAATGGCAGCTTGTTATAAAGAGAATACACCAAAAAGTTTAGAATATGCAACAAAACTATGGTCAGATATGGCTTCAACGGGAACATTACCACTCAGAAGAGCGGCGACTTCTTTTGCTGGACTCGCATTAAAACAGGGAGCTCCTCACATCGCCTTGGAATCAATATCAGGCCAACGACAGCATTATATCACAATTAGAAATATTAAGGCCATGGCTCTAGCTGATATGGGCAGAGTGGACGACGCAATACCGATATTACGAAGTGTATTAAACATCGATTCACCCGATCAGAAAGACAAGCACACATTCTTTGAAGAAACTATATCCAAAGTTGGTGAAGCAGTTGAAAAAACGAACAACAAGGATACCAAGATCGAATTCGAGGATGTCAGGAAGGCACTTATAGAGAGAGGACTCATTGATAAGGATACTCTCGACAGTCATCTGAATTTTGAAATTAAAATACAAAACAAGTTTAAGGATGCACCACGTGGAATGCCTTTTCAATCGAGAAACATGCCGTTTGGCCAACGAAAAATTAGGAAGGATTTGCAATAG
Protein
MSLFLNYFLKTSFSCNQRLFLPCRQLHSTQVRLLYAPATIGIDGYLQARKNVKEQFVNFSEKFKTKMNDFVSDSKNMIFTEDLKNMVHLAEPSDLQLVLNMIKKFNTQSTEFRFGSFVFGPVVMRMFHFLDAPKEALQCFEEPANDGFFDQQVSYQILLDLLYCHEMFDEMYRVFEKVQKKQINMTKFPKYCVVLIMAACYKENTPKSLEYATKLWSDMASTGTLPLRRAATSFAGLALKQGAPHIALESISGQRQHYITIRNIKAMALADMGRVDDAIPILRSVLNIDSPDQKDKHTFFEETISKVGEAVEKTNNKDTKIEFEDVRKALIERGLIDKDTLDSHLNFEIKIQNKFKDAPRGMPFQSRNMPFGQRKIRKDLQ
Summary
Uniprot
A0A1E1WQC4
A0A2A4JAC2
A0A212EPM3
A0A2H1VPG8
A0A194QWL5
A0A194QLB7
+ More
S4PXH8 A0A2W1B804 H9J3G9 A0A1B6JLF5 A0A1W4WVN5 A0A1B6FI95 A0A0K8T303 A0A0A9WKK6 A0A224XF09 A0A067QGI5 A0A069DTD2 A0A0P4VPB5 T1HT83 A0A2J7RDT2 A0A0V0G918 A0A023F0Z9 A0A226EUF2 A0A1B6DXB7 A0A1D2N898 A0A336MAL7 A0A336N2D2 A0A1Y1NBV0 D6WRX9 A0A2R7WAR2 J3JY18 N6U324 J9K1Z9 A0A2S2R467 U5ES26 A0A1J1I689 A0A2P8Z3Z6 A0A084VC54 A0A182ILS0 A0A151I7I2 E9IM06 A0A2M3ZA96 A0A3R7MFZ2 A0A195EAE9 A0A2M3ZA17 E2AYS9 W5JM20 A0A182PDN2 A0A2M4BQI9 A0A182THC6 A0A0N8DUZ7 A0A182I2Q1 A0A0P6H6X5 A0A0P4ZQA4 A0A0P6E8Z6 A0A182XLI5 A0A182KQM3 A0A0P5JFD9 Q5TWS3 A0A158NC65 A0A182K165 A0A0P5URY5 A0A2M4AV97 A0A0P6BHR5 A0A2M4AVE7 V5IA53 E9FQX6 A0A0P4ZKV1 A0A0P5MPQ5 A0A151I0M0 A0A0P6D7Z4 A0A0P5N848 A0A151XCR2 A0A182Q427 A0A182FGK4 A0A2M4BRI2 A0A182YMK6 A0A1B0DDZ8 Q16FZ9 Q16W31 A0A026WT99 A0A0P5N8R6 A0A182T0H6 A0A195F9A3 L7LYX6 A0A182N927 A0A0P5MR49 A0A1E1XK52 A0A2R5LG79 A0A3L8DFD8 A0A131XIW9 A0A182VFL6 A0A182R3J8 A0A1E1X9B7 A0A0P5TZ69 A0A1L8E0B6 A0A023FVP6
S4PXH8 A0A2W1B804 H9J3G9 A0A1B6JLF5 A0A1W4WVN5 A0A1B6FI95 A0A0K8T303 A0A0A9WKK6 A0A224XF09 A0A067QGI5 A0A069DTD2 A0A0P4VPB5 T1HT83 A0A2J7RDT2 A0A0V0G918 A0A023F0Z9 A0A226EUF2 A0A1B6DXB7 A0A1D2N898 A0A336MAL7 A0A336N2D2 A0A1Y1NBV0 D6WRX9 A0A2R7WAR2 J3JY18 N6U324 J9K1Z9 A0A2S2R467 U5ES26 A0A1J1I689 A0A2P8Z3Z6 A0A084VC54 A0A182ILS0 A0A151I7I2 E9IM06 A0A2M3ZA96 A0A3R7MFZ2 A0A195EAE9 A0A2M3ZA17 E2AYS9 W5JM20 A0A182PDN2 A0A2M4BQI9 A0A182THC6 A0A0N8DUZ7 A0A182I2Q1 A0A0P6H6X5 A0A0P4ZQA4 A0A0P6E8Z6 A0A182XLI5 A0A182KQM3 A0A0P5JFD9 Q5TWS3 A0A158NC65 A0A182K165 A0A0P5URY5 A0A2M4AV97 A0A0P6BHR5 A0A2M4AVE7 V5IA53 E9FQX6 A0A0P4ZKV1 A0A0P5MPQ5 A0A151I0M0 A0A0P6D7Z4 A0A0P5N848 A0A151XCR2 A0A182Q427 A0A182FGK4 A0A2M4BRI2 A0A182YMK6 A0A1B0DDZ8 Q16FZ9 Q16W31 A0A026WT99 A0A0P5N8R6 A0A182T0H6 A0A195F9A3 L7LYX6 A0A182N927 A0A0P5MR49 A0A1E1XK52 A0A2R5LG79 A0A3L8DFD8 A0A131XIW9 A0A182VFL6 A0A182R3J8 A0A1E1X9B7 A0A0P5TZ69 A0A1L8E0B6 A0A023FVP6
Pubmed
22118469
26354079
23622113
28756777
19121390
25401762
+ More
26823975 24845553 26334808 27129103 25474469 27289101 28004739 18362917 19820115 22516182 23537049 29403074 24438588 21282665 20798317 20920257 23761445 20966253 12364791 14747013 17210077 21347285 21292972 25244985 17510324 24508170 25576852 29209593 30249741 28049606 28503490
26823975 24845553 26334808 27129103 25474469 27289101 28004739 18362917 19820115 22516182 23537049 29403074 24438588 21282665 20798317 20920257 23761445 20966253 12364791 14747013 17210077 21347285 21292972 25244985 17510324 24508170 25576852 29209593 30249741 28049606 28503490
EMBL
GDQN01002008
JAT89046.1
NWSH01002390
PCG68390.1
AGBW02013432
OWR43455.1
+ More
ODYU01003670 SOQ42729.1 KQ461073 KPJ09699.1 KQ458575 KPJ05730.1 GAIX01005013 JAA87547.1 KZ150235 PZC71979.1 BABH01021397 GECU01007717 JAS99989.1 GECZ01019841 JAS49928.1 GBRD01006286 JAG59535.1 GBHO01034627 GDHC01021733 GDHC01008683 JAG08977.1 JAP96895.1 JAQ09946.1 GFTR01005359 JAW11067.1 KK853518 KDR07029.1 GBGD01001982 JAC86907.1 GDKW01002408 JAI54187.1 ACPB03016679 NEVH01005284 PNF38995.1 GECL01001662 JAP04462.1 GBBI01003547 JAC15165.1 LNIX01000001 OXA61243.1 GEDC01006982 JAS30316.1 LJIJ01000150 ODN01474.1 UFQT01000794 SSX27296.1 UFQS01004709 UFQT01004709 SSX16556.1 SSX35759.1 GEZM01007696 JAV94988.1 KQ971351 EFA06421.2 KK854531 PTY16608.1 BT128141 AEE63102.1 APGK01051641 KB741198 KB632313 ENN72972.1 ERL92168.1 ABLF02033333 ABLF02033339 GGMS01015555 MBY84758.1 GANO01003438 JAB56433.1 CVRI01000042 CRK95707.1 PYGN01000206 PSN51224.1 ATLV01010510 KE524585 KFB35548.1 KQ978417 KYM94058.1 GL764129 EFZ18479.1 GGFM01004703 MBW25454.1 QCYY01001094 ROT80662.1 KQ979236 KYN22195.1 GGFM01004636 MBW25387.1 GL443983 EFN61442.1 ADMH02001146 ETN63935.1 GGFJ01006141 MBW55282.1 GDIQ01089292 JAN05445.1 APCN01000178 GDIQ01024379 JAN70358.1 GDIP01210288 JAJ13114.1 GDIQ01077909 JAN16828.1 GDIQ01200415 GDIQ01197767 GDIQ01179311 GDIQ01083017 GDIQ01083016 GDIQ01067565 GDIQ01047474 LRGB01000642 JAK51310.1 KZS17306.1 AAAB01008807 EAL41847.2 ADTU01011668 ADTU01011669 GDIP01109350 JAL94364.1 GGFK01011374 MBW44695.1 GDIP01014382 JAM89333.1 GGFK01011448 MBW44769.1 GALX01001785 JAB66681.1 GL732523 EFX90291.1 GDIP01224435 JAI98966.1 GDIQ01153181 JAK98544.1 KQ976642 KYM78664.1 GDIQ01089291 JAN05446.1 GDIQ01153182 JAK98543.1 KQ982298 KYQ58166.1 AXCN02000540 GGFJ01006554 MBW55695.1 AJVK01032412 CH478354 EAT33163.1 CH477576 EAT38791.1 KK107109 EZA59237.1 GDIQ01149390 JAL02336.1 KQ981727 KYN37180.1 GACK01007753 JAA57281.1 GDIQ01153180 JAK98545.1 GFAA01003737 JAT99697.1 GGLE01004386 MBY08512.1 QOIP01000008 RLU19175.1 GEFH01002114 JAP66467.1 GFAC01003562 JAT95626.1 GDIP01122301 JAL81413.1 GFDF01001943 JAV12141.1 GBBL01002530 JAC24790.1
ODYU01003670 SOQ42729.1 KQ461073 KPJ09699.1 KQ458575 KPJ05730.1 GAIX01005013 JAA87547.1 KZ150235 PZC71979.1 BABH01021397 GECU01007717 JAS99989.1 GECZ01019841 JAS49928.1 GBRD01006286 JAG59535.1 GBHO01034627 GDHC01021733 GDHC01008683 JAG08977.1 JAP96895.1 JAQ09946.1 GFTR01005359 JAW11067.1 KK853518 KDR07029.1 GBGD01001982 JAC86907.1 GDKW01002408 JAI54187.1 ACPB03016679 NEVH01005284 PNF38995.1 GECL01001662 JAP04462.1 GBBI01003547 JAC15165.1 LNIX01000001 OXA61243.1 GEDC01006982 JAS30316.1 LJIJ01000150 ODN01474.1 UFQT01000794 SSX27296.1 UFQS01004709 UFQT01004709 SSX16556.1 SSX35759.1 GEZM01007696 JAV94988.1 KQ971351 EFA06421.2 KK854531 PTY16608.1 BT128141 AEE63102.1 APGK01051641 KB741198 KB632313 ENN72972.1 ERL92168.1 ABLF02033333 ABLF02033339 GGMS01015555 MBY84758.1 GANO01003438 JAB56433.1 CVRI01000042 CRK95707.1 PYGN01000206 PSN51224.1 ATLV01010510 KE524585 KFB35548.1 KQ978417 KYM94058.1 GL764129 EFZ18479.1 GGFM01004703 MBW25454.1 QCYY01001094 ROT80662.1 KQ979236 KYN22195.1 GGFM01004636 MBW25387.1 GL443983 EFN61442.1 ADMH02001146 ETN63935.1 GGFJ01006141 MBW55282.1 GDIQ01089292 JAN05445.1 APCN01000178 GDIQ01024379 JAN70358.1 GDIP01210288 JAJ13114.1 GDIQ01077909 JAN16828.1 GDIQ01200415 GDIQ01197767 GDIQ01179311 GDIQ01083017 GDIQ01083016 GDIQ01067565 GDIQ01047474 LRGB01000642 JAK51310.1 KZS17306.1 AAAB01008807 EAL41847.2 ADTU01011668 ADTU01011669 GDIP01109350 JAL94364.1 GGFK01011374 MBW44695.1 GDIP01014382 JAM89333.1 GGFK01011448 MBW44769.1 GALX01001785 JAB66681.1 GL732523 EFX90291.1 GDIP01224435 JAI98966.1 GDIQ01153181 JAK98544.1 KQ976642 KYM78664.1 GDIQ01089291 JAN05446.1 GDIQ01153182 JAK98543.1 KQ982298 KYQ58166.1 AXCN02000540 GGFJ01006554 MBW55695.1 AJVK01032412 CH478354 EAT33163.1 CH477576 EAT38791.1 KK107109 EZA59237.1 GDIQ01149390 JAL02336.1 KQ981727 KYN37180.1 GACK01007753 JAA57281.1 GDIQ01153180 JAK98545.1 GFAA01003737 JAT99697.1 GGLE01004386 MBY08512.1 QOIP01000008 RLU19175.1 GEFH01002114 JAP66467.1 GFAC01003562 JAT95626.1 GDIP01122301 JAL81413.1 GFDF01001943 JAV12141.1 GBBL01002530 JAC24790.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000005204
UP000192223
+ More
UP000027135 UP000015103 UP000235965 UP000198287 UP000094527 UP000007266 UP000019118 UP000030742 UP000007819 UP000183832 UP000245037 UP000030765 UP000075880 UP000078542 UP000283509 UP000078492 UP000000311 UP000000673 UP000075885 UP000075902 UP000075840 UP000076407 UP000075882 UP000076858 UP000007062 UP000005205 UP000075881 UP000000305 UP000078540 UP000075809 UP000075886 UP000069272 UP000076408 UP000092462 UP000008820 UP000053097 UP000075901 UP000078541 UP000075884 UP000279307 UP000075903 UP000075900
UP000027135 UP000015103 UP000235965 UP000198287 UP000094527 UP000007266 UP000019118 UP000030742 UP000007819 UP000183832 UP000245037 UP000030765 UP000075880 UP000078542 UP000283509 UP000078492 UP000000311 UP000000673 UP000075885 UP000075902 UP000075840 UP000076407 UP000075882 UP000076858 UP000007062 UP000005205 UP000075881 UP000000305 UP000078540 UP000075809 UP000075886 UP000069272 UP000076408 UP000092462 UP000008820 UP000053097 UP000075901 UP000078541 UP000075884 UP000279307 UP000075903 UP000075900
PRIDE
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A1E1WQC4
A0A2A4JAC2
A0A212EPM3
A0A2H1VPG8
A0A194QWL5
A0A194QLB7
+ More
S4PXH8 A0A2W1B804 H9J3G9 A0A1B6JLF5 A0A1W4WVN5 A0A1B6FI95 A0A0K8T303 A0A0A9WKK6 A0A224XF09 A0A067QGI5 A0A069DTD2 A0A0P4VPB5 T1HT83 A0A2J7RDT2 A0A0V0G918 A0A023F0Z9 A0A226EUF2 A0A1B6DXB7 A0A1D2N898 A0A336MAL7 A0A336N2D2 A0A1Y1NBV0 D6WRX9 A0A2R7WAR2 J3JY18 N6U324 J9K1Z9 A0A2S2R467 U5ES26 A0A1J1I689 A0A2P8Z3Z6 A0A084VC54 A0A182ILS0 A0A151I7I2 E9IM06 A0A2M3ZA96 A0A3R7MFZ2 A0A195EAE9 A0A2M3ZA17 E2AYS9 W5JM20 A0A182PDN2 A0A2M4BQI9 A0A182THC6 A0A0N8DUZ7 A0A182I2Q1 A0A0P6H6X5 A0A0P4ZQA4 A0A0P6E8Z6 A0A182XLI5 A0A182KQM3 A0A0P5JFD9 Q5TWS3 A0A158NC65 A0A182K165 A0A0P5URY5 A0A2M4AV97 A0A0P6BHR5 A0A2M4AVE7 V5IA53 E9FQX6 A0A0P4ZKV1 A0A0P5MPQ5 A0A151I0M0 A0A0P6D7Z4 A0A0P5N848 A0A151XCR2 A0A182Q427 A0A182FGK4 A0A2M4BRI2 A0A182YMK6 A0A1B0DDZ8 Q16FZ9 Q16W31 A0A026WT99 A0A0P5N8R6 A0A182T0H6 A0A195F9A3 L7LYX6 A0A182N927 A0A0P5MR49 A0A1E1XK52 A0A2R5LG79 A0A3L8DFD8 A0A131XIW9 A0A182VFL6 A0A182R3J8 A0A1E1X9B7 A0A0P5TZ69 A0A1L8E0B6 A0A023FVP6
S4PXH8 A0A2W1B804 H9J3G9 A0A1B6JLF5 A0A1W4WVN5 A0A1B6FI95 A0A0K8T303 A0A0A9WKK6 A0A224XF09 A0A067QGI5 A0A069DTD2 A0A0P4VPB5 T1HT83 A0A2J7RDT2 A0A0V0G918 A0A023F0Z9 A0A226EUF2 A0A1B6DXB7 A0A1D2N898 A0A336MAL7 A0A336N2D2 A0A1Y1NBV0 D6WRX9 A0A2R7WAR2 J3JY18 N6U324 J9K1Z9 A0A2S2R467 U5ES26 A0A1J1I689 A0A2P8Z3Z6 A0A084VC54 A0A182ILS0 A0A151I7I2 E9IM06 A0A2M3ZA96 A0A3R7MFZ2 A0A195EAE9 A0A2M3ZA17 E2AYS9 W5JM20 A0A182PDN2 A0A2M4BQI9 A0A182THC6 A0A0N8DUZ7 A0A182I2Q1 A0A0P6H6X5 A0A0P4ZQA4 A0A0P6E8Z6 A0A182XLI5 A0A182KQM3 A0A0P5JFD9 Q5TWS3 A0A158NC65 A0A182K165 A0A0P5URY5 A0A2M4AV97 A0A0P6BHR5 A0A2M4AVE7 V5IA53 E9FQX6 A0A0P4ZKV1 A0A0P5MPQ5 A0A151I0M0 A0A0P6D7Z4 A0A0P5N848 A0A151XCR2 A0A182Q427 A0A182FGK4 A0A2M4BRI2 A0A182YMK6 A0A1B0DDZ8 Q16FZ9 Q16W31 A0A026WT99 A0A0P5N8R6 A0A182T0H6 A0A195F9A3 L7LYX6 A0A182N927 A0A0P5MR49 A0A1E1XK52 A0A2R5LG79 A0A3L8DFD8 A0A131XIW9 A0A182VFL6 A0A182R3J8 A0A1E1X9B7 A0A0P5TZ69 A0A1L8E0B6 A0A023FVP6
Ontologies
PANTHER
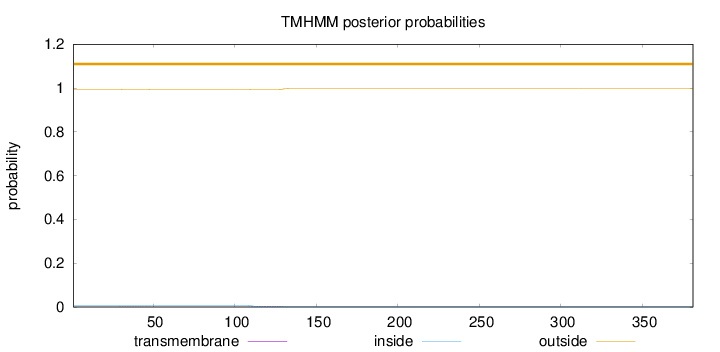
Topology
Length:
381
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0797899999999999
Exp number, first 60 AAs:
0.00472
Total prob of N-in:
0.00617
outside
1 - 381
Population Genetic Test Statistics
Pi
19.246866
Theta
157.768823
Tajima's D
-0.798628
CLR
0.470882
CSRT
0.172791360431978
Interpretation
Uncertain
