Gene
KWMTBOMO11507
Pre Gene Modal
BGIBMGA004057
Annotation
PREDICTED:_olfactory_receptor_49_isoform_X1_[Bombyx_mori]
Full name
Odorant receptor
Location in the cell
PlasmaMembrane Reliability : 4.779
Sequence
CDS
ATGATACGAACCCTTAAGACGGCTCGCAGCGCATCTTGGTTTGTCATAATCAATCAATCAGTGACCCATGTGATATATTTACTTATACCGTTCTTGTTCACCATTTTTGGGAATATCCGATATTTACCGACGACTCCCGGTGAAACTTATGGTTTGACTCCAAAATATGAGACTCCATATTACGAAATAACATTTATGCTAACATGTTTCGCGACAATATTCTCAGCCGTGAATCAGACAGGCTATATAGTGTTGTTCATCAACTTATTAGCTCACGAACTTGGACATTTTTATGTAATTACGGACGTCTTAAATGGAATTTTTGAAAAGAATGATGCCGATCGTGATCCTGTGTTTATAGATCGAAAATTGAAGTTCTGCGCGAAGCACTATCAATATTTACTAAAGTTCCATAACGAGATAAAAAACTTATATAAGATTATATTCGGCGCGCATTTCTTAATGATGACGATCGTTCTCGTGACGACATTACAGACGATGAATTCATGGGACATAAGGAATACGGTGCTTACAGCAGTTACAGGGATAATGCCCCTGTTCATATATTGTTTCGGGGGCGAATTGCTCATCACGGCGGGAATGGATATGTCTACAGCGATTTACCAGTGCGGGTGGGAGAAAATGGGCGTAAAACAAGCAAAGGTAGTCTCGGTTATCTTGTGTTTGTCTCAACGACCCTTATGTTTAACTGCGGCTAATGTTTTCGTTATGAATAGGGAGACATTCGGTGGTATCGCTCAGGTCGTTTACAAGATATACGCGGTATTTAACTAG
Protein
MIRTLKTARSASWFVIINQSVTHVIYLLIPFLFTIFGNIRYLPTTPGETYGLTPKYETPYYEITFMLTCFATIFSAVNQTGYIVLFINLLAHELGHFYVITDVLNGIFEKNDADRDPVFIDRKLKFCAKHYQYLLKFHNEIKNLYKIIFGAHFLMMTIVLVTTLQTMNSWDIRNTVLTAVTGIMPLFIYCFGGELLITAGMDMSTAIYQCGWEKMGVKQAKVVSVILCLSQRPLCLTAANVFVMNRETFGGIAQVVYKIYAVFN
Summary
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Uniprot
C4B7X7
A0A2H1WG60
A0A2A4J3B1
A0A2W1BDN1
A0A075T655
A0A3S2KZE4
+ More
A0A0L7KYU4 U5NJ08 A0A2K8GKV5 A0A194PE64 A0A2A4J488 A0A2K8GL41 A0A2W1BGJ8 A0A0M3SBN2 A0A0B5CMZ9 A0A0B5A1T0 A0A221I0J6 A0A0M4JNZ0 A0A0L7LUP7 B1B1Q4 A0A097IYL1 A0A0B5GI86 A0A223HD72 A0A2L1TG44 A0A223HDF1 C4B7Y2 A0A212ERN2 A0A0L7LUX5 A0A221I0E6 A0A194PKL6 C4B7Y3 A0A221I0I2 A0A1L2BLB8 A0A0M5K8H0 A0A2W1BIS7 A0A212FC10 A0A1S5XXN5 A0A1S5XXM9 A0A1Q1NIP3 A0A2P8Y155 A0A0M5K5R0 H9A5Q4 A0A212FKR6 A0A075T2Z3 A0A076E626 A0A0S1TRB2 A0A0M4J311 H9JAQ3 A0A1B3P5N6 A0A1X9P7J6 A0A146JWS3 A0A212F4V0 A0A0M4J7B6 A0A0K8TUT7 A0A2W1BH90 A0A0V0J1D8 A0A076E5Z5 A0A1Y9TJJ5 A0A386H9R7 A0A146JYU4 A0A2H1WCF2 I3Y3H7 B1B1Q7 A0A2K8GKS3 A0A221I0J4 A0A1S5XXN3 A0A1S5XXN2 H9JCV0 A0A2A4J308 A0A221I0A7 A0A2S2PJ41 A0A2L1TG55 A0A2A4JAT4 A0A1B3B746 A0A2P8YXB7 A0A2A4JBA0 A0A386H9B3 A0A151XJC6 A0A386H9N9 A0A0E3U2J7 A0A0M4JMM9 A0A1S5VFL2 A0A1V1WC68 A0A1W6L1F8 U5NJ75 A0A3L9LU14 A0A0L7K4E6 A0A151JBP1 A0A0K8TUS2 A0A075T642 A0A1S5XXQ0 A0A212FC05 E2BFJ9
A0A0L7KYU4 U5NJ08 A0A2K8GKV5 A0A194PE64 A0A2A4J488 A0A2K8GL41 A0A2W1BGJ8 A0A0M3SBN2 A0A0B5CMZ9 A0A0B5A1T0 A0A221I0J6 A0A0M4JNZ0 A0A0L7LUP7 B1B1Q4 A0A097IYL1 A0A0B5GI86 A0A223HD72 A0A2L1TG44 A0A223HDF1 C4B7Y2 A0A212ERN2 A0A0L7LUX5 A0A221I0E6 A0A194PKL6 C4B7Y3 A0A221I0I2 A0A1L2BLB8 A0A0M5K8H0 A0A2W1BIS7 A0A212FC10 A0A1S5XXN5 A0A1S5XXM9 A0A1Q1NIP3 A0A2P8Y155 A0A0M5K5R0 H9A5Q4 A0A212FKR6 A0A075T2Z3 A0A076E626 A0A0S1TRB2 A0A0M4J311 H9JAQ3 A0A1B3P5N6 A0A1X9P7J6 A0A146JWS3 A0A212F4V0 A0A0M4J7B6 A0A0K8TUT7 A0A2W1BH90 A0A0V0J1D8 A0A076E5Z5 A0A1Y9TJJ5 A0A386H9R7 A0A146JYU4 A0A2H1WCF2 I3Y3H7 B1B1Q7 A0A2K8GKS3 A0A221I0J4 A0A1S5XXN3 A0A1S5XXN2 H9JCV0 A0A2A4J308 A0A221I0A7 A0A2S2PJ41 A0A2L1TG55 A0A2A4JAT4 A0A1B3B746 A0A2P8YXB7 A0A2A4JBA0 A0A386H9B3 A0A151XJC6 A0A386H9N9 A0A0E3U2J7 A0A0M4JMM9 A0A1S5VFL2 A0A1V1WC68 A0A1W6L1F8 U5NJ75 A0A3L9LU14 A0A0L7K4E6 A0A151JBP1 A0A0K8TUS2 A0A075T642 A0A1S5XXQ0 A0A212FC05 E2BFJ9
Pubmed
EMBL
BABH01021396
AB472126
BAH66343.1
ODYU01008444
SOQ52063.1
NWSH01003519
+ More
PCG66178.1 KZ150235 PZC71965.1 KF768706 AIG51900.1 RSAL01000373 RVE42130.1 JTDY01004294 KOB68320.1 KC960472 AGY14583.1 KX585367 ARO70260.1 KQ459606 KPI91641.1 NWSH01003458 PCG66260.1 KY225496 ARO70508.1 KZ150062 PZC74192.1 KP843286 ALD51422.1 KM678312 AJE25885.1 KJ542684 AJD81568.1 KY964970 ASM47123.1 KP843355 ALD51491.1 JTDY01000047 KOB79193.1 AB186513 BAG12812.1 KM655549 AIT71999.1 KM892386 AJF23801.1 KY283662 AST36321.1 KY707206 AVF19648.1 KY283739 AST36398.1 AB472131 BAH66348.1 AGBW02012963 OWR44153.1 KOB79194.1 KY964983 ASM47136.1 KPI91640.1 AB472132 BAH66349.1 KY964950 ASM47103.1 KT860046 ALS03873.1 KP843249 ALD51385.1 PZC74191.1 AGBW02009241 OWR51272.1 KY399284 AQQ73515.1 KY399281 AQQ73512.1 KU958182 AQM56011.1 PYGN01001060 PSN37988.1 KP843237 ALD51373.1 JN836704 AFC91744.1 AGBW02008001 OWR54326.1 KF768704 AIG51898.1 KF487708 AII01106.1 KR935735 ALM26227.1 KP843344 ALD51480.1 BABH01008620 KX655979 AOG12928.1 KX084467 ARO76422.1 GEDO01000058 JAP88568.1 AGBW02010309 OWR48756.1 KP843278 ALD51414.1 GCVX01000194 JAI18036.1 PZC74198.1 GDKB01000050 JAP38446.1 KF487678 AII01076.1 KX084500 ARO76455.1 MG546636 AYD42259.1 GEDO01000057 JAP88569.1 ODYU01007660 SOQ50636.1 JQ794807 AFL70812.1 AB186516 AB472118 BAG12815.1 BAH66336.1 KX585345 ARO70238.1 KY964956 ASM47109.1 KY399285 AQQ73516.1 KY399282 AQQ73513.1 BABH01027717 BABH01027718 BABH01027719 BABH01027720 BABH01027721 PCG66259.1 KY964951 ASM47104.1 GGMR01016850 MBY29469.1 KY707207 AVF19649.1 NWSH01002174 PCG68969.1 KT588131 AOE48041.1 PYGN01000304 PSN48875.1 PCG68968.1 MG546620 AYD42243.1 KQ982074 KYQ60523.1 MG546606 AYD42229.1 KM251661 AKC58542.1 KP843363 ALD51499.1 KY445491 AQN78426.1 GENK01000073 JAV45840.1 KX609505 ARN17912.1 KC960478 AGY14589.1 KB466684 RLZ02265.1 JTDY01012011 KOB52347.1 KQ979175 KYN22365.1 GCVX01000209 JAI18021.1 KF768691 AIG51885.1 KY399296 AQQ73527.1 OWR51276.1 GL448037 EFN85521.1
PCG66178.1 KZ150235 PZC71965.1 KF768706 AIG51900.1 RSAL01000373 RVE42130.1 JTDY01004294 KOB68320.1 KC960472 AGY14583.1 KX585367 ARO70260.1 KQ459606 KPI91641.1 NWSH01003458 PCG66260.1 KY225496 ARO70508.1 KZ150062 PZC74192.1 KP843286 ALD51422.1 KM678312 AJE25885.1 KJ542684 AJD81568.1 KY964970 ASM47123.1 KP843355 ALD51491.1 JTDY01000047 KOB79193.1 AB186513 BAG12812.1 KM655549 AIT71999.1 KM892386 AJF23801.1 KY283662 AST36321.1 KY707206 AVF19648.1 KY283739 AST36398.1 AB472131 BAH66348.1 AGBW02012963 OWR44153.1 KOB79194.1 KY964983 ASM47136.1 KPI91640.1 AB472132 BAH66349.1 KY964950 ASM47103.1 KT860046 ALS03873.1 KP843249 ALD51385.1 PZC74191.1 AGBW02009241 OWR51272.1 KY399284 AQQ73515.1 KY399281 AQQ73512.1 KU958182 AQM56011.1 PYGN01001060 PSN37988.1 KP843237 ALD51373.1 JN836704 AFC91744.1 AGBW02008001 OWR54326.1 KF768704 AIG51898.1 KF487708 AII01106.1 KR935735 ALM26227.1 KP843344 ALD51480.1 BABH01008620 KX655979 AOG12928.1 KX084467 ARO76422.1 GEDO01000058 JAP88568.1 AGBW02010309 OWR48756.1 KP843278 ALD51414.1 GCVX01000194 JAI18036.1 PZC74198.1 GDKB01000050 JAP38446.1 KF487678 AII01076.1 KX084500 ARO76455.1 MG546636 AYD42259.1 GEDO01000057 JAP88569.1 ODYU01007660 SOQ50636.1 JQ794807 AFL70812.1 AB186516 AB472118 BAG12815.1 BAH66336.1 KX585345 ARO70238.1 KY964956 ASM47109.1 KY399285 AQQ73516.1 KY399282 AQQ73513.1 BABH01027717 BABH01027718 BABH01027719 BABH01027720 BABH01027721 PCG66259.1 KY964951 ASM47104.1 GGMR01016850 MBY29469.1 KY707207 AVF19649.1 NWSH01002174 PCG68969.1 KT588131 AOE48041.1 PYGN01000304 PSN48875.1 PCG68968.1 MG546620 AYD42243.1 KQ982074 KYQ60523.1 MG546606 AYD42229.1 KM251661 AKC58542.1 KP843363 ALD51499.1 KY445491 AQN78426.1 GENK01000073 JAV45840.1 KX609505 ARN17912.1 KC960478 AGY14589.1 KB466684 RLZ02265.1 JTDY01012011 KOB52347.1 KQ979175 KYN22365.1 GCVX01000209 JAI18021.1 KF768691 AIG51885.1 KY399296 AQQ73527.1 OWR51276.1 GL448037 EFN85521.1
Proteomes
Pfam
PF02949 7tm_6
Interpro
IPR004117
7tm6_olfct_rcpt
ProteinModelPortal
C4B7X7
A0A2H1WG60
A0A2A4J3B1
A0A2W1BDN1
A0A075T655
A0A3S2KZE4
+ More
A0A0L7KYU4 U5NJ08 A0A2K8GKV5 A0A194PE64 A0A2A4J488 A0A2K8GL41 A0A2W1BGJ8 A0A0M3SBN2 A0A0B5CMZ9 A0A0B5A1T0 A0A221I0J6 A0A0M4JNZ0 A0A0L7LUP7 B1B1Q4 A0A097IYL1 A0A0B5GI86 A0A223HD72 A0A2L1TG44 A0A223HDF1 C4B7Y2 A0A212ERN2 A0A0L7LUX5 A0A221I0E6 A0A194PKL6 C4B7Y3 A0A221I0I2 A0A1L2BLB8 A0A0M5K8H0 A0A2W1BIS7 A0A212FC10 A0A1S5XXN5 A0A1S5XXM9 A0A1Q1NIP3 A0A2P8Y155 A0A0M5K5R0 H9A5Q4 A0A212FKR6 A0A075T2Z3 A0A076E626 A0A0S1TRB2 A0A0M4J311 H9JAQ3 A0A1B3P5N6 A0A1X9P7J6 A0A146JWS3 A0A212F4V0 A0A0M4J7B6 A0A0K8TUT7 A0A2W1BH90 A0A0V0J1D8 A0A076E5Z5 A0A1Y9TJJ5 A0A386H9R7 A0A146JYU4 A0A2H1WCF2 I3Y3H7 B1B1Q7 A0A2K8GKS3 A0A221I0J4 A0A1S5XXN3 A0A1S5XXN2 H9JCV0 A0A2A4J308 A0A221I0A7 A0A2S2PJ41 A0A2L1TG55 A0A2A4JAT4 A0A1B3B746 A0A2P8YXB7 A0A2A4JBA0 A0A386H9B3 A0A151XJC6 A0A386H9N9 A0A0E3U2J7 A0A0M4JMM9 A0A1S5VFL2 A0A1V1WC68 A0A1W6L1F8 U5NJ75 A0A3L9LU14 A0A0L7K4E6 A0A151JBP1 A0A0K8TUS2 A0A075T642 A0A1S5XXQ0 A0A212FC05 E2BFJ9
A0A0L7KYU4 U5NJ08 A0A2K8GKV5 A0A194PE64 A0A2A4J488 A0A2K8GL41 A0A2W1BGJ8 A0A0M3SBN2 A0A0B5CMZ9 A0A0B5A1T0 A0A221I0J6 A0A0M4JNZ0 A0A0L7LUP7 B1B1Q4 A0A097IYL1 A0A0B5GI86 A0A223HD72 A0A2L1TG44 A0A223HDF1 C4B7Y2 A0A212ERN2 A0A0L7LUX5 A0A221I0E6 A0A194PKL6 C4B7Y3 A0A221I0I2 A0A1L2BLB8 A0A0M5K8H0 A0A2W1BIS7 A0A212FC10 A0A1S5XXN5 A0A1S5XXM9 A0A1Q1NIP3 A0A2P8Y155 A0A0M5K5R0 H9A5Q4 A0A212FKR6 A0A075T2Z3 A0A076E626 A0A0S1TRB2 A0A0M4J311 H9JAQ3 A0A1B3P5N6 A0A1X9P7J6 A0A146JWS3 A0A212F4V0 A0A0M4J7B6 A0A0K8TUT7 A0A2W1BH90 A0A0V0J1D8 A0A076E5Z5 A0A1Y9TJJ5 A0A386H9R7 A0A146JYU4 A0A2H1WCF2 I3Y3H7 B1B1Q7 A0A2K8GKS3 A0A221I0J4 A0A1S5XXN3 A0A1S5XXN2 H9JCV0 A0A2A4J308 A0A221I0A7 A0A2S2PJ41 A0A2L1TG55 A0A2A4JAT4 A0A1B3B746 A0A2P8YXB7 A0A2A4JBA0 A0A386H9B3 A0A151XJC6 A0A386H9N9 A0A0E3U2J7 A0A0M4JMM9 A0A1S5VFL2 A0A1V1WC68 A0A1W6L1F8 U5NJ75 A0A3L9LU14 A0A0L7K4E6 A0A151JBP1 A0A0K8TUS2 A0A075T642 A0A1S5XXQ0 A0A212FC05 E2BFJ9
Ontologies
GO
PANTHER
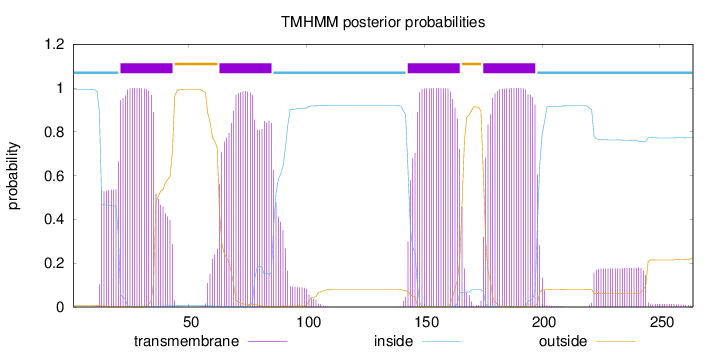
Topology
Subcellular location
Cell membrane
Length:
264
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
95.38123
Exp number, first 60 AAs:
23.20382
Total prob of N-in:
0.99338
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 62
TMhelix
63 - 85
inside
86 - 142
TMhelix
143 - 165
outside
166 - 174
TMhelix
175 - 197
inside
198 - 264
Population Genetic Test Statistics
Pi
179.079594
Theta
172.970991
Tajima's D
-0.760198
CLR
0.537014
CSRT
0.189090545472726
Interpretation
Uncertain
