Gene
KWMTBOMO11506
Pre Gene Modal
BGIBMGA001910
Annotation
PREDICTED:_diuretic_hormone_receptor_[Bombyx_mori]
Full name
Diuretic hormone receptor
Location in the cell
PlasmaMembrane Reliability : 4.967
Sequence
CDS
ATGACGGGAAAGTACTCGGCATTGATCACATCATACATTACAAACATGCTAATTACGTTGTACGGGCAAGAAATGTTAAATGACACCGCCATAATTGAAGCAAAAACGAAAGTTCCTTGTACGATACGCAAATACTTCTGCAGACTGCCGATGTTTACTTTCGGAGCGTTATCACTTGATAAGTTAGAACACAAGCCGCGTGTAAACATTTTTATCATGTACATTTTTAACGGCAAACTGGAGGACTTACAGACTTTAGAAAGCGCAGAAAGCGTTATCGAAAATGCGACTGCAAGGACCGAGTGCTTGGCGAGGAACGCTAATCTCACAGAATACTACTGCCCGGCATATTTTGATGGATTGTTGTGTTGGAACCCGACGCCGAGCCACACGGTCGCCGTCCAGAAATGCTTTAAAGAATTTTTTGGAATAAAATACGATGAAACACAAAACGCTTCTCGTCTGTGTCTGGACGGTGTTTGGCAGAACTACACCGACTACTCGAATTGCACGGAGCGGATCGCTAATGTGTCACCCACGGACGTGGCCAGTCTCATATACTTGACGGGATACTCTCTCAGTCTCGCCGTTTTATCGCTGGCCGTCTTCGTCTTCTTGTATTTTAAGGATCTAAGATGCTTAAGAAACACGATACATACAAATTTAATGACCACATATATACTATCGGCTTGTAGTTGGATATTAAATTTAGCTTTACAAAATTGGTCCGACGAGGCACAGCAAGATCAAACGTCATGCATGATCTTAGTTATATGCATGCATTATTTTTATCTAACGAATTTCTTCTGGATGCTCGTTGAAGGTCTGTACCTGTACATGCTTGTTGTGGAAACATTCACCGCGGAAAATATTAAACTCAAAGTATACACTACCATCGGTTGGGGTGCTCCGGCGATATTCATCACAATTTGGGTCGTGTCAAGATGTTTTGTCAACGTCATGCCATCGACTGGACCAGACGGTCTTGCGCTTGCTGGGGAAGCGAAGATGTGTATATGGATGCACGAACATCAGGTGGACTGGATACACAAAGCTCCCGCGTTGGCGGGACTCGCGCTGAACTTATTTTTCCTTGTCAGGATTATGTGGGTGCTGATTACGAAACTCCGCTCGGCGAACACCCTCGAAACAGAACAATACAGGAAGGCTACTAAAGCGCTTCTTGTATTGATACCATTGTTGGGTATCACTAACTTGCTTGTGCTCTGCGGACCTAGTGACGACTCCTGGTTCGCGTACTCATTCGACTACGCGAGAGCCCTCATGCTATCAACACAGGGATTCACAGTGGCCCTATTCTATTGCTTCATGAACACTGAGGTGAGACACGCGATCAGGTACCACGTTGAGAGATGGAAGACTGGCAGAAATATCGGAGGAGGGAGAAGGAGGGGAGCTTCATACTCTAAGGACTGGTCTCCTAGATCCAGAACGGAGAGTATACGGTTATACAACCACCCGTCGAAGCGAGAGTCTGCGGCTTCGGAGACGACCACCACCACACTCCTCGTGCCGAGGACCTCACTCGCACCGCACTATGACTCCAGGCACCTACACGCTGACTACATCACGAGCGGTAGCGCTAGCGGGGAACAGTCTCCGGTTTGA
Protein
MTGKYSALITSYITNMLITLYGQEMLNDTAIIEAKTKVPCTIRKYFCRLPMFTFGALSLDKLEHKPRVNIFIMYIFNGKLEDLQTLESAESVIENATARTECLARNANLTEYYCPAYFDGLLCWNPTPSHTVAVQKCFKEFFGIKYDETQNASRLCLDGVWQNYTDYSNCTERIANVSPTDVASLIYLTGYSLSLAVLSLAVFVFLYFKDLRCLRNTIHTNLMTTYILSACSWILNLALQNWSDEAQQDQTSCMILVICMHYFYLTNFFWMLVEGLYLYMLVVETFTAENIKLKVYTTIGWGAPAIFITIWVVSRCFVNVMPSTGPDGLALAGEAKMCIWMHEHQVDWIHKAPALAGLALNLFFLVRIMWVLITKLRSANTLETEQYRKATKALLVLIPLLGITNLLVLCGPSDDSWFAYSFDYARALMLSTQGFTVALFYCFMNTEVRHAIRYHVERWKTGRNIGGGRRRGASYSKDWSPRSRTESIRLYNHPSKRESAASETTTTTLLVPRTSLAPHYDSRHLHADYITSGSASGEQSPV
Summary
Similarity
Belongs to the G-protein coupled receptor 2 family.
Keywords
Cell membrane
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Transducer
Transmembrane
Transmembrane helix
Signal
Feature
chain Diuretic hormone receptor
Uniprot
A0A194QXL6
A0A194QK26
P35464
A0A0S1YDC3
G3CKA8
B6VCU7
+ More
B6VCU3 A0A1B6DBY6 Q65AS2 A0A1Y1KDD1 A0A139WBV2 A0A1Y1KJG5 Q65AS3 A0A1Y1KDE3 Q16983 A0A224XR65 A0A2U9PG90 B3FN73 B4F7L3 A0A1Q3FJD0 A0A1Q3FK63 A0A139WBU8 A0A1Q3FKA1 A0A1Q3FK13 A0A1Q3FK45 A0A182RFL3 A0A1Q3FK52 A0A2M4CI98 W5JKI3 A0A0A1WY41 A0A3B0J3W5 Q28X78 A0A2H8TNI8 A0A0R3NQ01 J9M7L7 A0A3B0JN39 A0A0R3NPR2 A0A0Q9W0T1 A0A1I8MWK2 A0A0M4ETT2 A0A182LYB7 A0A0Q9W896 A0A182Y0U9 A0A336LUX8 B3NRZ9 A0A2C9GPK6 Q7Q773 A0A0J9RAY4 B3MEN2 Q4V3E9 B4N601 B4HPJ8 B4QCX4 B4P592 A0A0Q9XG64 A0A034W5C9 A0A1W4UJB3 E0VEH3 A0A2M4CR10 A0A2M4CYV5 A0A0R1DW56 A0A084W2V1 A0A0K8UG33 B4J886 A0A1I8Q291 A0A1I8Q273 A0A3B0J0Z9 A0A1W4UHG8 A0A1I8MMV0 A0A0K8V0F2 A0A182UZ74 W8C899 A0A0L0C399 A0A0K0LFR2 A0A2C9GPP6 A0A034WB61 T1H976 A0A0K8U0V0 A0A0K8VM25 A0A0K8VJV2 A0A182FFC9 W8BFD2 A0A182Y0U8 A0A1B6CDW7 A0A182LJP0 A0A1B6GNH4 A0A182TZL7 A0A0Q5VMV0 A0A1B6IFI2
B6VCU3 A0A1B6DBY6 Q65AS2 A0A1Y1KDD1 A0A139WBV2 A0A1Y1KJG5 Q65AS3 A0A1Y1KDE3 Q16983 A0A224XR65 A0A2U9PG90 B3FN73 B4F7L3 A0A1Q3FJD0 A0A1Q3FK63 A0A139WBU8 A0A1Q3FKA1 A0A1Q3FK13 A0A1Q3FK45 A0A182RFL3 A0A1Q3FK52 A0A2M4CI98 W5JKI3 A0A0A1WY41 A0A3B0J3W5 Q28X78 A0A2H8TNI8 A0A0R3NQ01 J9M7L7 A0A3B0JN39 A0A0R3NPR2 A0A0Q9W0T1 A0A1I8MWK2 A0A0M4ETT2 A0A182LYB7 A0A0Q9W896 A0A182Y0U9 A0A336LUX8 B3NRZ9 A0A2C9GPK6 Q7Q773 A0A0J9RAY4 B3MEN2 Q4V3E9 B4N601 B4HPJ8 B4QCX4 B4P592 A0A0Q9XG64 A0A034W5C9 A0A1W4UJB3 E0VEH3 A0A2M4CR10 A0A2M4CYV5 A0A0R1DW56 A0A084W2V1 A0A0K8UG33 B4J886 A0A1I8Q291 A0A1I8Q273 A0A3B0J0Z9 A0A1W4UHG8 A0A1I8MMV0 A0A0K8V0F2 A0A182UZ74 W8C899 A0A0L0C399 A0A0K0LFR2 A0A2C9GPP6 A0A034WB61 T1H976 A0A0K8U0V0 A0A0K8VM25 A0A0K8VJV2 A0A182FFC9 W8BFD2 A0A182Y0U8 A0A1B6CDW7 A0A182LJP0 A0A1B6GNH4 A0A182TZL7 A0A0Q5VMV0 A0A1B6IFI2
Pubmed
26354079
8276884
22093064
15373805
28004739
18362917
+ More
19820115 8673074 18651923 20920257 23761445 25830018 15632085 17994087 23185243 25315136 18057021 25244985 12364791 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 20566863 24438588 24495485 26108605 20966253
19820115 8673074 18651923 20920257 23761445 25830018 15632085 17994087 23185243 25315136 18057021 25244985 12364791 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 20566863 24438588 24495485 26108605 20966253
EMBL
KQ461073
KPJ09700.1
KQ458575
KPJ05729.1
U03489
KT031047
+ More
ALM88345.1 HQ008729 AEK82133.1 FJ374694 ACJ06654.1 FJ374690 ACJ06650.1 GEDC01014104 JAS23194.1 AJ634781 CAG25576.1 GEZM01086343 GEZM01086341 JAV59532.1 KQ971372 KYB25406.1 GEZM01086338 GEZM01086336 JAV59536.1 AJ634780 CAG25575.2 GEZM01086342 GEZM01086340 JAV59533.1 U15959 GFTR01005925 JAW10501.1 MG550231 AWT50665.1 EU273352 ABX57920.1 BK006347 DAA06284.1 GFDL01007314 JAV27731.1 GFDL01007162 JAV27883.1 KYB25407.1 GFDL01007132 JAV27913.1 GFDL01007136 JAV27909.1 GFDL01007172 JAV27873.1 GFDL01007173 JAV27872.1 GGFL01000791 MBW64969.1 ADMH02000928 ETN64646.1 GBXI01011004 JAD03288.1 OUUW01000001 SPP74053.1 CM000071 EAL26438.2 GFXV01003765 MBW15570.1 KRT03106.1 ABLF02041105 ABLF02041114 ABLF02041126 ABLF02041133 ABLF02041157 SPP74051.1 KRT03100.1 KRT03102.1 KRT03104.1 CH940662 KRF78422.1 CP012524 ALC40695.1 AXCM01005703 AXCM01005704 KRF78420.1 KRF78421.1 KRF78423.1 KRF78424.1 KRF78425.1 KRF78426.1 KRF78427.1 UFQS01000007 UFQT01000007 SSW97072.1 SSX17458.1 CH954179 EDV56301.1 APCN01004347 APCN01004348 APCN01004349 AAAB01008960 EAA11768.4 CM002911 KMY93223.1 CH902619 EDV35496.2 KPU75698.1 KPU75699.1 KPU75701.1 AE013599 BT023407 AAF58501.4 AAY55823.1 CH964154 EDW79790.2 CH480816 EDW47582.1 CM000362 EDX06785.1 KMY93218.1 CM000158 EDW90754.1 CH933808 KRG03906.1 GAKP01009019 GAKP01009018 JAC49934.1 DS235090 EEB11779.1 GGFL01003150 MBW67328.1 GGFL01006281 MBW70459.1 KRJ99447.1 KRJ99448.1 KRJ99449.1 ATLV01019701 ATLV01019702 ATLV01019703 KE525278 KFB44545.1 GDHF01026677 GDHF01026131 JAI25637.1 JAI26183.1 CH916367 EDW01223.1 SPP74744.1 GDHF01019930 JAI32384.1 GAMC01007611 JAB98944.1 JRES01000960 KNC26741.1 KJ407397 AJA04995.1 APCN01004353 GAKP01006161 JAC52791.1 ACPB03016148 ACPB03016149 GDHF01031987 JAI20327.1 GDHF01012704 JAI39610.1 GDHF01013150 JAI39164.1 GAMC01010907 JAB95648.1 GEDC01025652 JAS11646.1 GECZ01005773 JAS63996.1 KQS62832.1 GECU01028762 GECU01022031 JAS78944.1 JAS85675.1
ALM88345.1 HQ008729 AEK82133.1 FJ374694 ACJ06654.1 FJ374690 ACJ06650.1 GEDC01014104 JAS23194.1 AJ634781 CAG25576.1 GEZM01086343 GEZM01086341 JAV59532.1 KQ971372 KYB25406.1 GEZM01086338 GEZM01086336 JAV59536.1 AJ634780 CAG25575.2 GEZM01086342 GEZM01086340 JAV59533.1 U15959 GFTR01005925 JAW10501.1 MG550231 AWT50665.1 EU273352 ABX57920.1 BK006347 DAA06284.1 GFDL01007314 JAV27731.1 GFDL01007162 JAV27883.1 KYB25407.1 GFDL01007132 JAV27913.1 GFDL01007136 JAV27909.1 GFDL01007172 JAV27873.1 GFDL01007173 JAV27872.1 GGFL01000791 MBW64969.1 ADMH02000928 ETN64646.1 GBXI01011004 JAD03288.1 OUUW01000001 SPP74053.1 CM000071 EAL26438.2 GFXV01003765 MBW15570.1 KRT03106.1 ABLF02041105 ABLF02041114 ABLF02041126 ABLF02041133 ABLF02041157 SPP74051.1 KRT03100.1 KRT03102.1 KRT03104.1 CH940662 KRF78422.1 CP012524 ALC40695.1 AXCM01005703 AXCM01005704 KRF78420.1 KRF78421.1 KRF78423.1 KRF78424.1 KRF78425.1 KRF78426.1 KRF78427.1 UFQS01000007 UFQT01000007 SSW97072.1 SSX17458.1 CH954179 EDV56301.1 APCN01004347 APCN01004348 APCN01004349 AAAB01008960 EAA11768.4 CM002911 KMY93223.1 CH902619 EDV35496.2 KPU75698.1 KPU75699.1 KPU75701.1 AE013599 BT023407 AAF58501.4 AAY55823.1 CH964154 EDW79790.2 CH480816 EDW47582.1 CM000362 EDX06785.1 KMY93218.1 CM000158 EDW90754.1 CH933808 KRG03906.1 GAKP01009019 GAKP01009018 JAC49934.1 DS235090 EEB11779.1 GGFL01003150 MBW67328.1 GGFL01006281 MBW70459.1 KRJ99447.1 KRJ99448.1 KRJ99449.1 ATLV01019701 ATLV01019702 ATLV01019703 KE525278 KFB44545.1 GDHF01026677 GDHF01026131 JAI25637.1 JAI26183.1 CH916367 EDW01223.1 SPP74744.1 GDHF01019930 JAI32384.1 GAMC01007611 JAB98944.1 JRES01000960 KNC26741.1 KJ407397 AJA04995.1 APCN01004353 GAKP01006161 JAC52791.1 ACPB03016148 ACPB03016149 GDHF01031987 JAI20327.1 GDHF01012704 JAI39610.1 GDHF01013150 JAI39164.1 GAMC01010907 JAB95648.1 GEDC01025652 JAS11646.1 GECZ01005773 JAS63996.1 KQS62832.1 GECU01028762 GECU01022031 JAS78944.1 JAS85675.1
Proteomes
UP000053240
UP000053268
UP000007266
UP000075900
UP000000673
UP000268350
+ More
UP000001819 UP000007819 UP000008792 UP000095301 UP000092553 UP000075883 UP000076408 UP000008711 UP000075840 UP000007062 UP000007801 UP000000803 UP000007798 UP000001292 UP000000304 UP000002282 UP000009192 UP000192221 UP000009046 UP000030765 UP000001070 UP000095300 UP000075903 UP000037069 UP000015103 UP000069272 UP000075882 UP000075902
UP000001819 UP000007819 UP000008792 UP000095301 UP000092553 UP000075883 UP000076408 UP000008711 UP000075840 UP000007062 UP000007801 UP000000803 UP000007798 UP000001292 UP000000304 UP000002282 UP000009192 UP000192221 UP000009046 UP000030765 UP000001070 UP000095300 UP000075903 UP000037069 UP000015103 UP000069272 UP000075882 UP000075902
PRIDE
Interpro
SUPFAM
SSF111418
SSF111418
Gene 3D
ProteinModelPortal
A0A194QXL6
A0A194QK26
P35464
A0A0S1YDC3
G3CKA8
B6VCU7
+ More
B6VCU3 A0A1B6DBY6 Q65AS2 A0A1Y1KDD1 A0A139WBV2 A0A1Y1KJG5 Q65AS3 A0A1Y1KDE3 Q16983 A0A224XR65 A0A2U9PG90 B3FN73 B4F7L3 A0A1Q3FJD0 A0A1Q3FK63 A0A139WBU8 A0A1Q3FKA1 A0A1Q3FK13 A0A1Q3FK45 A0A182RFL3 A0A1Q3FK52 A0A2M4CI98 W5JKI3 A0A0A1WY41 A0A3B0J3W5 Q28X78 A0A2H8TNI8 A0A0R3NQ01 J9M7L7 A0A3B0JN39 A0A0R3NPR2 A0A0Q9W0T1 A0A1I8MWK2 A0A0M4ETT2 A0A182LYB7 A0A0Q9W896 A0A182Y0U9 A0A336LUX8 B3NRZ9 A0A2C9GPK6 Q7Q773 A0A0J9RAY4 B3MEN2 Q4V3E9 B4N601 B4HPJ8 B4QCX4 B4P592 A0A0Q9XG64 A0A034W5C9 A0A1W4UJB3 E0VEH3 A0A2M4CR10 A0A2M4CYV5 A0A0R1DW56 A0A084W2V1 A0A0K8UG33 B4J886 A0A1I8Q291 A0A1I8Q273 A0A3B0J0Z9 A0A1W4UHG8 A0A1I8MMV0 A0A0K8V0F2 A0A182UZ74 W8C899 A0A0L0C399 A0A0K0LFR2 A0A2C9GPP6 A0A034WB61 T1H976 A0A0K8U0V0 A0A0K8VM25 A0A0K8VJV2 A0A182FFC9 W8BFD2 A0A182Y0U8 A0A1B6CDW7 A0A182LJP0 A0A1B6GNH4 A0A182TZL7 A0A0Q5VMV0 A0A1B6IFI2
B6VCU3 A0A1B6DBY6 Q65AS2 A0A1Y1KDD1 A0A139WBV2 A0A1Y1KJG5 Q65AS3 A0A1Y1KDE3 Q16983 A0A224XR65 A0A2U9PG90 B3FN73 B4F7L3 A0A1Q3FJD0 A0A1Q3FK63 A0A139WBU8 A0A1Q3FKA1 A0A1Q3FK13 A0A1Q3FK45 A0A182RFL3 A0A1Q3FK52 A0A2M4CI98 W5JKI3 A0A0A1WY41 A0A3B0J3W5 Q28X78 A0A2H8TNI8 A0A0R3NQ01 J9M7L7 A0A3B0JN39 A0A0R3NPR2 A0A0Q9W0T1 A0A1I8MWK2 A0A0M4ETT2 A0A182LYB7 A0A0Q9W896 A0A182Y0U9 A0A336LUX8 B3NRZ9 A0A2C9GPK6 Q7Q773 A0A0J9RAY4 B3MEN2 Q4V3E9 B4N601 B4HPJ8 B4QCX4 B4P592 A0A0Q9XG64 A0A034W5C9 A0A1W4UJB3 E0VEH3 A0A2M4CR10 A0A2M4CYV5 A0A0R1DW56 A0A084W2V1 A0A0K8UG33 B4J886 A0A1I8Q291 A0A1I8Q273 A0A3B0J0Z9 A0A1W4UHG8 A0A1I8MMV0 A0A0K8V0F2 A0A182UZ74 W8C899 A0A0L0C399 A0A0K0LFR2 A0A2C9GPP6 A0A034WB61 T1H976 A0A0K8U0V0 A0A0K8VM25 A0A0K8VJV2 A0A182FFC9 W8BFD2 A0A182Y0U8 A0A1B6CDW7 A0A182LJP0 A0A1B6GNH4 A0A182TZL7 A0A0Q5VMV0 A0A1B6IFI2
PDB
5UZ7
E-value=2.04163e-32,
Score=349
Ontologies
KEGG
GO
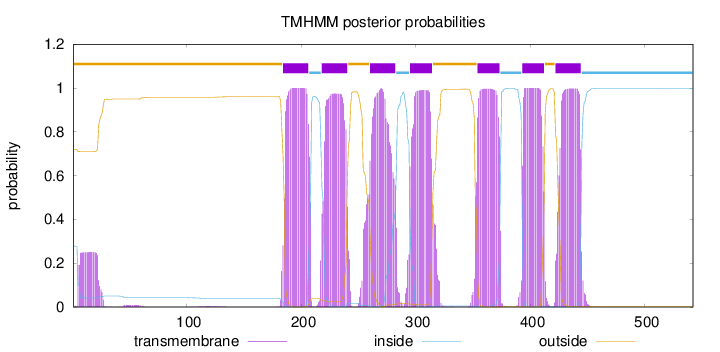
Topology
Subcellular location
Cell membrane
Length:
542
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
153.30553
Exp number, first 60 AAs:
4.91283
Total prob of N-in:
0.28096
outside
1 - 183
TMhelix
184 - 206
inside
207 - 217
TMhelix
218 - 240
outside
241 - 259
TMhelix
260 - 282
inside
283 - 294
TMhelix
295 - 314
outside
315 - 353
TMhelix
354 - 373
inside
374 - 392
TMhelix
393 - 412
outside
413 - 421
TMhelix
422 - 444
inside
445 - 542
Population Genetic Test Statistics
Pi
201.107883
Theta
172.531651
Tajima's D
0.794386
CLR
0.164627
CSRT
0.603769811509425
Interpretation
Uncertain
