Gene
KWMTBOMO11502
Annotation
PREDICTED:_uncharacterized_protein_LOC106136784_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.873
Sequence
CDS
ATGAATACATTCATGTCGGGGGAGCTGCACACGTTCACCGACGCCAGCGAGAAGGCCTACGCGTGTGCAGTCTATTGGCGTCAGAAGGCAGACGAGAGCACCTACCGCGTCACGCTTCTAGCAGGGAAAGCCAGGGTGACCCCACTGAGACCAGTCTCTATCCCACGGCTGGAGCTGCAAGCTGCGCTGCTGGGGACAAGGATGGCGCAGGCGATAGCGGACGAATTGGACATCGCAGTTGGTGTAAGGACGTACTGGACTGACTCCAGTACAGTCCTGACATGGATAAAGACCGACCCGCGCACGTTCAAACCTTTCGTTGCGCACCGACTCGCCGAGATAGAAGAGTCAACGAAGCCCCAAGAATGGCGATGGGTGCCTGGCTCGCAAAACCCAGCAGACGACGCAACTAGAGAGGCGCCGGCGGATTTCGACCACACGCATCGGTGGTTTAATGGACCCGAGTTCCTGACGTGGGATGAATCGCGCTGGCCCAAGCCGCGAAAATTCAAGCAAGAACCGTCTGGAGAAGAGAAAGAGGCCTACCTGGTCGCTACGGCGAGGACCGCTGACGCTCAACCCACGCCCGATCCGCACAGATTCTCGAGCTGGGTCAGGTTATTGAGAGCAACAGCCAGGGTCCTCCAATTTATCGAGCTGTGTCGACCCCAGAAGGAGAGCGCCTGCGTGTCCAGACAATCGAAGCAGCAAGATCCCACGTGGAGGACGACACGAACGAAGCAACCCCGATCCACATGGAAGTTAAGGACCCCGGAAGCACCTACAGAAGCATGGCTGCCGCTTGATCCGCCCCACTTGAAGAAGGCGGAGAAGATCCTACTGAGAAGCAGCCAAAGAGAGAGCTTCGGTGAAAAATATCCCGAACGTCACCCCAAGTTGCGACGGCTCGACGTCGTGATGGAAGATGGCCTGTTGCGCCTGCGCGGACGTATCGACGCAGCGCAATACATAGATGCAGGCTGCAAGCGACCGATCGTGCTGGACGGGAAGCACGTGATAGCGAGATTATTGATCAAACACTATCACGAGGCGTTCCAGCACGTGGTGCAAGGTCTATCGGAGTACACCACGACTACCGCCCACCGGATACCTGCCGATAGAGCGGCTACGACATGGAGAGCCGCCGTTCACCTGTGCTGCCGTCGATTACTTCGGGCCGATGACGGTGACTGTAGGACGACGTCACGAGAAGAGGTGGGGCGTGCTGTTCACATGCCTCACCACGAGGCAGACTCAATGCTGTTAGCGCTGCGCAGGATGGCCGCCCGACGAGGAATGCCCAAAGTCATTTACAGCGACAATGGGACTAACTTCGTCGGCGCCAACAAGGAGCTCAAGCAGGCAATCGAGAATGCAAGGGAAGCAGACGTCGTGTCACGGGCCGCTCAGATGAACATAAAATGGAAGTTCATCCCGCCAGGAGCGCCGAACATGGGAGGCGCGTGGGAACGTTTGGTGCGGTCCGTGAAGACCGCGCTGGCGGTCACACTGAAGGAAAGACACCCGAGGGAAGAAGTTCTGCACACTCTTCTGTTGGAGGCGGAGCATGTCGTCAACTCGAGACCGCTCGTCGCTAGGGAAGAGAGTTGGGAGTCAGAAGCACTCACGCCGAACGACTTTCTCATAGGGAGATCCTGCGGAGCTCCGAGCATCGGCGACTACAGAGACGAAGACCTGACCGGCAAAAGGACATGGAGAGTGGCGCAGCGTATGGCGGACCACTTCTGGAGTAGGTGGGTCAAGGAATACCTGCCTACGCTGCTGCCCAGGAAGATCGACGGGAGAGCAGCTGGCGAGGACTTGCAGTGTGGGGATACCGTCCTCATAGTGGACTCCACGCTGCCACGCAACACATGGCCGCGCGGGCAAGTTGTCCACACCTACCCAGGCCCCGACCACCGTGTACGAACTGTCGACGTCAGAACGAGCGGCGGTCTGCTTAAGAGACCTGCCTCCAAGATGATCCTGCTGGTGCCTGCAACGAGCGACGAGCCCACGCCCATAGAGTCGCCACGACAAGGAGTCGGTGCTACGCACGAGGGGGAGGATGTTGGCGACAGCGGCGTCGCCCCATAA
Protein
MNTFMSGELHTFTDASEKAYACAVYWRQKADESTYRVTLLAGKARVTPLRPVSIPRLELQAALLGTRMAQAIADELDIAVGVRTYWTDSSTVLTWIKTDPRTFKPFVAHRLAEIEESTKPQEWRWVPGSQNPADDATREAPADFDHTHRWFNGPEFLTWDESRWPKPRKFKQEPSGEEKEAYLVATARTADAQPTPDPHRFSSWVRLLRATARVLQFIELCRPQKESACVSRQSKQQDPTWRTTRTKQPRSTWKLRTPEAPTEAWLPLDPPHLKKAEKILLRSSQRESFGEKYPERHPKLRRLDVVMEDGLLRLRGRIDAAQYIDAGCKRPIVLDGKHVIARLLIKHYHEAFQHVVQGLSEYTTTTAHRIPADRAATTWRAAVHLCCRRLLRADDGDCRTTSREEVGRAVHMPHHEADSMLLALRRMAARRGMPKVIYSDNGTNFVGANKELKQAIENAREADVVSRAAQMNIKWKFIPPGAPNMGGAWERLVRSVKTALAVTLKERHPREEVLHTLLLEAEHVVNSRPLVAREESWESEALTPNDFLIGRSCGAPSIGDYRDEDLTGKRTWRVAQRMADHFWSRWVKEYLPTLLPRKIDGRAAGEDLQCGDTVLIVDSTLPRNTWPRGQVVHTYPGPDHRVRTVDVRTSGGLLKRPASKMILLVPATSDEPTPIESPRQGVGATHEGEDVGDSGVAP
Summary
Uniprot
Q2MGA5
A0A3S2N5J7
A0A0N1IGH3
K7JA41
A0A226DGY5
A0A226D351
+ More
A0A226EDU9 A0A226DH22 A0A226E2P7 A0A226EEI4 A0A226DM06 A0A226EFV7 A0A182GD64 W8AJC8 A0A182HDR0 A0A085NF60 A0A085MRE0 A0A0N5E596 A0A034V9M7 W4ZKP7 W4XEJ9 A0A182H5B8 A0A085NQS3 A0A182GRX9 A0A146QJ75 W4Y2R6 A0A2B4RJC8 A0A1B0CTR4 W4Z131 W4XEJ5 A0A182G1E0 A0A2B4SX75 W4XIU3 W4ZDM8 W4XIU5 A0A1W7R6D5 A0A085MU62 W4XIU6 A0A146TEM1 W4XIU4 A0A085MSJ2 A0A146T9I5 A0A3S2P6R4 A0A1S3K952 A0A1S3IZI0 A0A1S3IN25 W4Z2K2 A0A1S3I2B0 A0A1A8MUQ1 W4XIU2 W4Y4R6 W4YQI7 W4Y4R7 A0A2B4RBG6 W4ZBS6 A0A1A8U834 I3KZ01 A0A146SEB9 A0A085NCZ0 A0A0S7GQ49 A0A1A8V6P1 W4ZCQ1 A0A085ND08 A0A0V1NG14 A0A0V1NDW9 A0A0V1DCQ2 A0A2G8KEP2 A0A1S3IV31 A0A2B4RT98 W4XEJ6 W4XEK1 A0A1B0D7Y5 A0A1B0D864 A0A146PQY4 A0A085LRU1 A0A182GED3 A0A182VT46 A0A182HER3
A0A226EDU9 A0A226DH22 A0A226E2P7 A0A226EEI4 A0A226DM06 A0A226EFV7 A0A182GD64 W8AJC8 A0A182HDR0 A0A085NF60 A0A085MRE0 A0A0N5E596 A0A034V9M7 W4ZKP7 W4XEJ9 A0A182H5B8 A0A085NQS3 A0A182GRX9 A0A146QJ75 W4Y2R6 A0A2B4RJC8 A0A1B0CTR4 W4Z131 W4XEJ5 A0A182G1E0 A0A2B4SX75 W4XIU3 W4ZDM8 W4XIU5 A0A1W7R6D5 A0A085MU62 W4XIU6 A0A146TEM1 W4XIU4 A0A085MSJ2 A0A146T9I5 A0A3S2P6R4 A0A1S3K952 A0A1S3IZI0 A0A1S3IN25 W4Z2K2 A0A1S3I2B0 A0A1A8MUQ1 W4XIU2 W4Y4R6 W4YQI7 W4Y4R7 A0A2B4RBG6 W4ZBS6 A0A1A8U834 I3KZ01 A0A146SEB9 A0A085NCZ0 A0A0S7GQ49 A0A1A8V6P1 W4ZCQ1 A0A085ND08 A0A0V1NG14 A0A0V1NDW9 A0A0V1DCQ2 A0A2G8KEP2 A0A1S3IV31 A0A2B4RT98 W4XEJ6 W4XEK1 A0A1B0D7Y5 A0A1B0D864 A0A146PQY4 A0A085LRU1 A0A182GED3 A0A182VT46 A0A182HER3
EMBL
AF530470
AAQ09229.1
RSAL01000375
RVE42106.1
KQ459986
KPJ18861.1
+ More
AAZX01023278 LNIX01000019 OXA44812.1 LNIX01000039 OXA39294.1 LNIX01000004 OXA55408.1 OXA44855.1 LNIX01000007 OXA52015.1 OXA55241.1 LNIX01000015 OXA46575.1 OXA56250.1 JXUM01009840 KQ560293 KXJ83223.1 GAMC01017930 JAB88625.1 JXUM01130316 KQ567562 KXJ69370.1 KL367508 KFD68106.1 KL367737 KFD59786.1 GAKP01020709 JAC38243.1 AAGJ04095911 AAGJ04115911 JXUM01111221 KQ565482 KXJ70922.1 KL367480 KFD71819.1 JXUM01083542 KQ563383 KXJ73941.1 GCES01130050 JAQ56272.1 AAGJ04043389 LSMT01000554 PFX16342.1 AJWK01027898 AAGJ04040339 JXUM01038372 KQ561162 KXJ79503.1 LSMT01000016 PFX33015.1 AAGJ04001285 AAGJ04011066 GEHC01000921 JAV46724.1 KL367650 KFD60758.1 GCES01095024 JAQ91298.1 AAGJ04091218 KL367687 KFD60188.1 GCES01096813 JAQ89509.1 RSAL01000010 RVE53755.1 AAGJ04152028 HAEF01019456 SBR60615.1 AAGJ04173907 AAGJ04146728 AAGJ04146729 AAGJ04141734 LSMT01000699 PFX15001.1 AAGJ04122915 HAEJ01004007 SBS44464.1 AERX01020956 GCES01107353 JAQ78969.1 KL367515 KFD67336.1 GBYX01453073 JAO28442.1 HAEJ01014775 SBS55232.1 AAGJ04162434 KFD67354.1 JYDM01000241 KRZ82930.1 JYDM01000403 KRZ82016.1 JYDI01000018 KRY58771.1 MRZV01000643 PIK46464.1 LSMT01000268 PFX21664.1 AJVK01033431 AJVK01037041 GCES01140316 JAQ46006.1 KL363318 KFD47687.1 JXUM01162450 KQ574455 KXJ67869.1 JXUM01005801 KQ560202 KXJ83866.1
AAZX01023278 LNIX01000019 OXA44812.1 LNIX01000039 OXA39294.1 LNIX01000004 OXA55408.1 OXA44855.1 LNIX01000007 OXA52015.1 OXA55241.1 LNIX01000015 OXA46575.1 OXA56250.1 JXUM01009840 KQ560293 KXJ83223.1 GAMC01017930 JAB88625.1 JXUM01130316 KQ567562 KXJ69370.1 KL367508 KFD68106.1 KL367737 KFD59786.1 GAKP01020709 JAC38243.1 AAGJ04095911 AAGJ04115911 JXUM01111221 KQ565482 KXJ70922.1 KL367480 KFD71819.1 JXUM01083542 KQ563383 KXJ73941.1 GCES01130050 JAQ56272.1 AAGJ04043389 LSMT01000554 PFX16342.1 AJWK01027898 AAGJ04040339 JXUM01038372 KQ561162 KXJ79503.1 LSMT01000016 PFX33015.1 AAGJ04001285 AAGJ04011066 GEHC01000921 JAV46724.1 KL367650 KFD60758.1 GCES01095024 JAQ91298.1 AAGJ04091218 KL367687 KFD60188.1 GCES01096813 JAQ89509.1 RSAL01000010 RVE53755.1 AAGJ04152028 HAEF01019456 SBR60615.1 AAGJ04173907 AAGJ04146728 AAGJ04146729 AAGJ04141734 LSMT01000699 PFX15001.1 AAGJ04122915 HAEJ01004007 SBS44464.1 AERX01020956 GCES01107353 JAQ78969.1 KL367515 KFD67336.1 GBYX01453073 JAO28442.1 HAEJ01014775 SBS55232.1 AAGJ04162434 KFD67354.1 JYDM01000241 KRZ82930.1 JYDM01000403 KRZ82016.1 JYDI01000018 KRY58771.1 MRZV01000643 PIK46464.1 LSMT01000268 PFX21664.1 AJVK01033431 AJVK01037041 GCES01140316 JAQ46006.1 KL363318 KFD47687.1 JXUM01162450 KQ574455 KXJ67869.1 JXUM01005801 KQ560202 KXJ83866.1
Proteomes
Pfam
Interpro
IPR040676
DUF5641
+ More
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR041588 Integrase_H2C2
IPR008737 Peptidase_asp_put
IPR001995 Peptidase_A2_cat
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR001841 Znf_RING
IPR019786 Zinc_finger_PHD-type_CS
IPR000477 RT_dom
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR009563 SSSCA1
IPR036305 RGS_sf
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR000239 GPCR_kinase
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR016137 RGS
IPR036875 Znf_CCHC_sf
IPR032675 LRR_dom_sf
IPR027417 P-loop_NTPase
IPR007111 NACHT_NTPase
IPR001611 Leu-rich_rpt
IPR041249 HEPN_DZIP3
IPR013087 Znf_C2H2_type
IPR006594 LisH
IPR006595 CTLH_C
IPR024964 CTLH/CRA
IPR021109 Peptidase_aspartic_dom_sf
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR041588 Integrase_H2C2
IPR008737 Peptidase_asp_put
IPR001995 Peptidase_A2_cat
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR001841 Znf_RING
IPR019786 Zinc_finger_PHD-type_CS
IPR000477 RT_dom
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR009563 SSSCA1
IPR036305 RGS_sf
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR000239 GPCR_kinase
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR016137 RGS
IPR036875 Znf_CCHC_sf
IPR032675 LRR_dom_sf
IPR027417 P-loop_NTPase
IPR007111 NACHT_NTPase
IPR001611 Leu-rich_rpt
IPR041249 HEPN_DZIP3
IPR013087 Znf_C2H2_type
IPR006594 LisH
IPR006595 CTLH_C
IPR024964 CTLH/CRA
IPR021109 Peptidase_aspartic_dom_sf
SUPFAM
ProteinModelPortal
Q2MGA5
A0A3S2N5J7
A0A0N1IGH3
K7JA41
A0A226DGY5
A0A226D351
+ More
A0A226EDU9 A0A226DH22 A0A226E2P7 A0A226EEI4 A0A226DM06 A0A226EFV7 A0A182GD64 W8AJC8 A0A182HDR0 A0A085NF60 A0A085MRE0 A0A0N5E596 A0A034V9M7 W4ZKP7 W4XEJ9 A0A182H5B8 A0A085NQS3 A0A182GRX9 A0A146QJ75 W4Y2R6 A0A2B4RJC8 A0A1B0CTR4 W4Z131 W4XEJ5 A0A182G1E0 A0A2B4SX75 W4XIU3 W4ZDM8 W4XIU5 A0A1W7R6D5 A0A085MU62 W4XIU6 A0A146TEM1 W4XIU4 A0A085MSJ2 A0A146T9I5 A0A3S2P6R4 A0A1S3K952 A0A1S3IZI0 A0A1S3IN25 W4Z2K2 A0A1S3I2B0 A0A1A8MUQ1 W4XIU2 W4Y4R6 W4YQI7 W4Y4R7 A0A2B4RBG6 W4ZBS6 A0A1A8U834 I3KZ01 A0A146SEB9 A0A085NCZ0 A0A0S7GQ49 A0A1A8V6P1 W4ZCQ1 A0A085ND08 A0A0V1NG14 A0A0V1NDW9 A0A0V1DCQ2 A0A2G8KEP2 A0A1S3IV31 A0A2B4RT98 W4XEJ6 W4XEK1 A0A1B0D7Y5 A0A1B0D864 A0A146PQY4 A0A085LRU1 A0A182GED3 A0A182VT46 A0A182HER3
A0A226EDU9 A0A226DH22 A0A226E2P7 A0A226EEI4 A0A226DM06 A0A226EFV7 A0A182GD64 W8AJC8 A0A182HDR0 A0A085NF60 A0A085MRE0 A0A0N5E596 A0A034V9M7 W4ZKP7 W4XEJ9 A0A182H5B8 A0A085NQS3 A0A182GRX9 A0A146QJ75 W4Y2R6 A0A2B4RJC8 A0A1B0CTR4 W4Z131 W4XEJ5 A0A182G1E0 A0A2B4SX75 W4XIU3 W4ZDM8 W4XIU5 A0A1W7R6D5 A0A085MU62 W4XIU6 A0A146TEM1 W4XIU4 A0A085MSJ2 A0A146T9I5 A0A3S2P6R4 A0A1S3K952 A0A1S3IZI0 A0A1S3IN25 W4Z2K2 A0A1S3I2B0 A0A1A8MUQ1 W4XIU2 W4Y4R6 W4YQI7 W4Y4R7 A0A2B4RBG6 W4ZBS6 A0A1A8U834 I3KZ01 A0A146SEB9 A0A085NCZ0 A0A0S7GQ49 A0A1A8V6P1 W4ZCQ1 A0A085ND08 A0A0V1NG14 A0A0V1NDW9 A0A0V1DCQ2 A0A2G8KEP2 A0A1S3IV31 A0A2B4RT98 W4XEJ6 W4XEK1 A0A1B0D7Y5 A0A1B0D864 A0A146PQY4 A0A085LRU1 A0A182GED3 A0A182VT46 A0A182HER3
Ontologies
GO
Topology
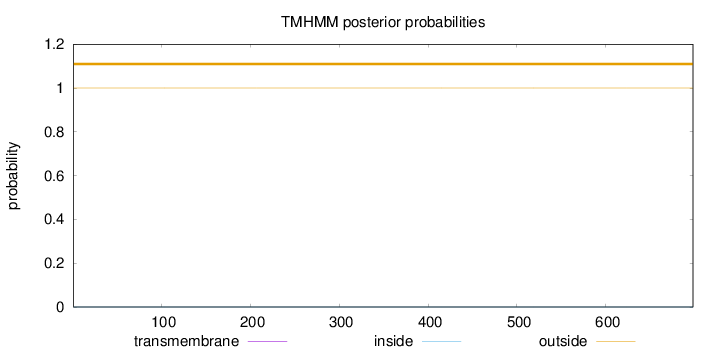
Length:
698
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00008
outside
1 - 698
Population Genetic Test Statistics
Pi
279.659573
Theta
171.922653
Tajima's D
1.985575
CLR
0.605367
CSRT
0.88025598720064
Interpretation
Uncertain
