Gene
KWMTBOMO11493
Annotation
PREDICTED:_papilin-like_[Amyelois_transitella]
Full name
Papilin
Location in the cell
Nuclear Reliability : 2.919
Sequence
CDS
ATGAACGCACAATCGACGAGCACCGAAATCGGACGACCGATATCGAGAACGACAGATGTATTACGAACGAACACGACGCTGTACGCGTGGAGTTCCTGGGGCAGGTGGAGTCCTTGCTCGAGGAGCTGCGGCGGAGGAGTTGCTGTACAAGAACGCCATTGCCTGCCTAGAATTCCTTCGAGAACGCGTCGCAGTCGAAAAAGAAGGAAGCTAACGAGAAACTTCGAGCACAAAAGACGGTCTCGGCATTTGATCCACGGCCCTGCCACAGAGCCGGATTGTCCAGGCCTCGCGCGGAGGTACCACGAGTGCAATACTGCGCCATGCTCGGGACCACAACGAGACGTTCGAGCCGAGCAATGCGCTGTCTACGATCGCAGACCGTTCCGAGGTCGATTCTATACTTGGGTGCCATATGTCGATGGAGACGCGCCATGTACCTTGAATTGTAGACCTCGTGGCCAGCAGTTCTACGCTTCCCTCGCTCTAGTTGCTGACGGCACCCCGTGTACTAAGCCAGGCTTTAGAGCAATATGTGTACAGGGAACATGCAAGACACACTAA
Protein
MNAQSTSTEIGRPISRTTDVLRTNTTLYAWSSWGRWSPCSRSCGGGVAVQERHCLPRIPSRTRRSRKRRKLTRNFEHKRRSRHLIHGPATEPDCPGLARRYHECNTAPCSGPQRDVRAEQCAVYDRRPFRGRFYTWVPYVDGDAPCTLNCRPRGQQFYASLALVADGTPCTKPGFRAICVQGTCKTH
Summary
Similarity
Belongs to the papilin family.
Keywords
Complete proteome
Disulfide bond
Immunoglobulin domain
Protease inhibitor
Reference proteome
Repeat
Secreted
Serine protease inhibitor
Signal
Feature
chain Papilin
Uniprot
A0A2A4K788
A0A194QXM2
A0A194QK21
A0A212FNQ0
A0A194QVZ7
A0A194QLA7
+ More
A0A2A4K7Z3 A0A212EYQ0 A0A1Y1MD44 A0A1Y1M948 A0A034WK37 A0A0K8UP59 W8BJ09 A0A0R3NW20 A0A0J7L0V3 A0A1B6JFE9 A0A154PM38 A0A182F546 A0A2M3ZC89 A0A2M3ZCE1 A0A1I8PD91 B4KHR2 B4JQ02 F7BTW4 A0A139WEE8 B4LVB9 F6YPW6 A0A087ZTP2 A0A0L7RBC8 E2B927 Q7Q128 K7FL94 A0A0C9S2T5 K7FLD6 A0A195D6Q8 B4Q9A9 B4HWQ2 Q29R29 Q9VKV3 A0A151I0E7 E1JHE3 A0A0N7ZCV3 A0A0P4WFN0 B4P0P5 A0A310SHM2 A0A3B3QJJ2 B4MUB3 Q8MSF8 A0A091PJB2 A0A3B3QKE1 A0A158NJN3 A0A182PW42 A0A3L8DX48 B3N5F4 D3ZD40 A0A026W198 A0A386H774 A0A3L8SXQ7 H0ZLV6 A0A195FPF2 A0A3Q3KQK2 L8Y4A4 A0A091MGK3 G3SMX7 A0A3B3CPB9 H0X0C8 A0A093DQZ5 A0A182JER1 A0A1U7QTI5 A0A091TDY8 A0A1U7QQV9 A0A3B3CP76 Q9EPX2 B7ZN28 A0A093IU08 A0A3B3DZ05 A0A340XID0 A0A2J8PJB5 A0A2R9CCA8 A0A2R9CCH0 H2R995 A0A2I3SKU7 A0A2J8T4F8 A0A2J8T4D1 H2NLQ3 A0A2J8T4D6 A0A195EKD7 A0A2I3T2D6 A0A2J8PJD7 A0A096NVY9 A0A2K5NMV5 A0A2J8PJD5 A0A2K5NN26 A0A2J8T4C5 A0A2K5NMV9 A0A2K6TCZ1 A0A2K6TCW2 A0A1B6M8Z1
A0A2A4K7Z3 A0A212EYQ0 A0A1Y1MD44 A0A1Y1M948 A0A034WK37 A0A0K8UP59 W8BJ09 A0A0R3NW20 A0A0J7L0V3 A0A1B6JFE9 A0A154PM38 A0A182F546 A0A2M3ZC89 A0A2M3ZCE1 A0A1I8PD91 B4KHR2 B4JQ02 F7BTW4 A0A139WEE8 B4LVB9 F6YPW6 A0A087ZTP2 A0A0L7RBC8 E2B927 Q7Q128 K7FL94 A0A0C9S2T5 K7FLD6 A0A195D6Q8 B4Q9A9 B4HWQ2 Q29R29 Q9VKV3 A0A151I0E7 E1JHE3 A0A0N7ZCV3 A0A0P4WFN0 B4P0P5 A0A310SHM2 A0A3B3QJJ2 B4MUB3 Q8MSF8 A0A091PJB2 A0A3B3QKE1 A0A158NJN3 A0A182PW42 A0A3L8DX48 B3N5F4 D3ZD40 A0A026W198 A0A386H774 A0A3L8SXQ7 H0ZLV6 A0A195FPF2 A0A3Q3KQK2 L8Y4A4 A0A091MGK3 G3SMX7 A0A3B3CPB9 H0X0C8 A0A093DQZ5 A0A182JER1 A0A1U7QTI5 A0A091TDY8 A0A1U7QQV9 A0A3B3CP76 Q9EPX2 B7ZN28 A0A093IU08 A0A3B3DZ05 A0A340XID0 A0A2J8PJB5 A0A2R9CCA8 A0A2R9CCH0 H2R995 A0A2I3SKU7 A0A2J8T4F8 A0A2J8T4D1 H2NLQ3 A0A2J8T4D6 A0A195EKD7 A0A2I3T2D6 A0A2J8PJD7 A0A096NVY9 A0A2K5NMV5 A0A2J8PJD5 A0A2K5NN26 A0A2J8T4C5 A0A2K5NMV9 A0A2K6TCZ1 A0A2K6TCW2 A0A1B6M8Z1
Pubmed
26354079
22118469
28004739
25348373
24495485
15632085
+ More
17994087 17495919 18362917 19820115 18057021 20431018 20798317 12364791 17381049 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 29240929 21347285 30249741 15057822 24508170 30282656 20360741 23385571 29451363 11076767 15489334 19468303 22722832 16136131
17994087 17495919 18362917 19820115 18057021 20431018 20798317 12364791 17381049 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 29240929 21347285 30249741 15057822 24508170 30282656 20360741 23385571 29451363 11076767 15489334 19468303 22722832 16136131
EMBL
NWSH01000085
PCG79774.1
KQ461073
KPJ09705.1
KQ458575
KPJ05724.1
+ More
AGBW02004790 OWR55371.1 KPJ09708.1 KPJ05720.1 PCG79780.1 AGBW02011484 OWR46574.1 GEZM01037051 JAV82225.1 GEZM01037053 JAV82224.1 GAKP01003001 JAC55951.1 GDHF01023948 JAI28366.1 GAMC01007823 JAB98732.1 CH379058 KRT03551.1 LBMM01001469 KMQ96261.1 GECU01010133 JAS97573.1 KQ434977 KZC12893.1 GGFM01005370 MBW26121.1 GGFM01005369 MBW26120.1 CH933807 EDW13349.2 CH916372 EDV98982.1 KQ971354 KYB26304.1 CH940649 EDW63298.2 KRF81024.1 KRF81025.1 AAMC01110600 KQ414617 KOC68227.1 GL446425 EFN87793.1 AAAB01008980 EAA14143.3 AGCU01123411 AGCU01123412 AGCU01123413 AGCU01123414 AGCU01123415 AGCU01123416 GBYB01014270 GBYB01015211 JAG84037.1 JAG84978.1 KQ976818 KYN08109.1 CM000361 CM002910 EDX04551.1 KMY89550.1 CH480818 EDW52447.1 BT024211 ABC86273.1 AE014134 AAF52956.3 KQ976666 KYM78288.1 ACZ94227.2 GDRN01059605 JAI65488.1 GDRN01059606 JAI65487.1 CM000157 EDW87940.1 KQ768636 OAD53080.1 CH963857 EDW76039.2 AY118849 AAM50709.1 KL390934 KFP91590.1 ADTU01018081 ADTU01018082 ADTU01018083 ADTU01018084 QOIP01000003 RLU24509.1 CH954177 EDV59033.1 AABR07065081 KK107499 EZA49788.1 MG452690 AYD41577.1 QUSF01000004 RLW10538.1 ABQF01002505 ABQF01002506 ABQF01002507 ABQF01002508 ABQF01002509 KQ981424 KYN41824.1 KB369403 ELV09874.1 KK827240 KFP73678.1 AAQR03188716 AAQR03188717 AAQR03188718 AAQR03188719 AAQR03188720 AAQR03188721 KL229237 KFV04566.1 KK949358 KFQ57036.1 AF314171 BC005747 BC132475 BC137854 AC132954 BC144975 AAI44976.1 KK569056 KFW06215.1 NBAG03000214 PNI84119.1 AJFE02084882 AJFE02084883 AACZ04008424 AACZ04008425 AACZ04008426 PNI84115.1 PNI84118.1 NDHI03003521 PNJ27886.1 PNJ27885.1 ABGA01343809 ABGA01343810 PNJ27880.1 KQ978782 KYN28387.1 PNI84116.1 AHZZ02032394 PNI84117.1 PNJ27884.1 GEBQ01007574 JAT32403.1
AGBW02004790 OWR55371.1 KPJ09708.1 KPJ05720.1 PCG79780.1 AGBW02011484 OWR46574.1 GEZM01037051 JAV82225.1 GEZM01037053 JAV82224.1 GAKP01003001 JAC55951.1 GDHF01023948 JAI28366.1 GAMC01007823 JAB98732.1 CH379058 KRT03551.1 LBMM01001469 KMQ96261.1 GECU01010133 JAS97573.1 KQ434977 KZC12893.1 GGFM01005370 MBW26121.1 GGFM01005369 MBW26120.1 CH933807 EDW13349.2 CH916372 EDV98982.1 KQ971354 KYB26304.1 CH940649 EDW63298.2 KRF81024.1 KRF81025.1 AAMC01110600 KQ414617 KOC68227.1 GL446425 EFN87793.1 AAAB01008980 EAA14143.3 AGCU01123411 AGCU01123412 AGCU01123413 AGCU01123414 AGCU01123415 AGCU01123416 GBYB01014270 GBYB01015211 JAG84037.1 JAG84978.1 KQ976818 KYN08109.1 CM000361 CM002910 EDX04551.1 KMY89550.1 CH480818 EDW52447.1 BT024211 ABC86273.1 AE014134 AAF52956.3 KQ976666 KYM78288.1 ACZ94227.2 GDRN01059605 JAI65488.1 GDRN01059606 JAI65487.1 CM000157 EDW87940.1 KQ768636 OAD53080.1 CH963857 EDW76039.2 AY118849 AAM50709.1 KL390934 KFP91590.1 ADTU01018081 ADTU01018082 ADTU01018083 ADTU01018084 QOIP01000003 RLU24509.1 CH954177 EDV59033.1 AABR07065081 KK107499 EZA49788.1 MG452690 AYD41577.1 QUSF01000004 RLW10538.1 ABQF01002505 ABQF01002506 ABQF01002507 ABQF01002508 ABQF01002509 KQ981424 KYN41824.1 KB369403 ELV09874.1 KK827240 KFP73678.1 AAQR03188716 AAQR03188717 AAQR03188718 AAQR03188719 AAQR03188720 AAQR03188721 KL229237 KFV04566.1 KK949358 KFQ57036.1 AF314171 BC005747 BC132475 BC137854 AC132954 BC144975 AAI44976.1 KK569056 KFW06215.1 NBAG03000214 PNI84119.1 AJFE02084882 AJFE02084883 AACZ04008424 AACZ04008425 AACZ04008426 PNI84115.1 PNI84118.1 NDHI03003521 PNJ27886.1 PNJ27885.1 ABGA01343809 ABGA01343810 PNJ27880.1 KQ978782 KYN28387.1 PNI84116.1 AHZZ02032394 PNI84117.1 PNJ27884.1 GEBQ01007574 JAT32403.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000001819
UP000036403
+ More
UP000076502 UP000069272 UP000095300 UP000009192 UP000001070 UP000002280 UP000007266 UP000008792 UP000008143 UP000005203 UP000053825 UP000008237 UP000007062 UP000007267 UP000078542 UP000000304 UP000001292 UP000000803 UP000078540 UP000002282 UP000261540 UP000007798 UP000005205 UP000075885 UP000279307 UP000008711 UP000002494 UP000053097 UP000276834 UP000007754 UP000078541 UP000261600 UP000011518 UP000007646 UP000261560 UP000005225 UP000075880 UP000189706 UP000000589 UP000265300 UP000240080 UP000002277 UP000001595 UP000078492 UP000028761 UP000233060 UP000233220
UP000076502 UP000069272 UP000095300 UP000009192 UP000001070 UP000002280 UP000007266 UP000008792 UP000008143 UP000005203 UP000053825 UP000008237 UP000007062 UP000007267 UP000078542 UP000000304 UP000001292 UP000000803 UP000078540 UP000002282 UP000261540 UP000007798 UP000005205 UP000075885 UP000279307 UP000008711 UP000002494 UP000053097 UP000276834 UP000007754 UP000078541 UP000261600 UP000011518 UP000007646 UP000261560 UP000005225 UP000075880 UP000189706 UP000000589 UP000265300 UP000240080 UP000002277 UP000001595 UP000078492 UP000028761 UP000233060 UP000233220
Pfam
Interpro
IPR036383
TSP1_rpt_sf
+ More
IPR000884 TSP1_rpt
IPR013273 ADAMTS/ADAMTS-like
IPR010294 ADAM_spacer1
IPR010909 PLAC
IPR003582 ShKT_dom
IPR036880 Kunitz_BPTI_sf
IPR020901 Prtase_inh_Kunz-CS
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR002223 Kunitz_BPTI
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR013106 Ig_V-set
IPR013151 Immunoglobulin
IPR000884 TSP1_rpt
IPR013273 ADAMTS/ADAMTS-like
IPR010294 ADAM_spacer1
IPR010909 PLAC
IPR003582 ShKT_dom
IPR036880 Kunitz_BPTI_sf
IPR020901 Prtase_inh_Kunz-CS
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR002223 Kunitz_BPTI
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR013106 Ig_V-set
IPR013151 Immunoglobulin
Gene 3D
CDD
ProteinModelPortal
A0A2A4K788
A0A194QXM2
A0A194QK21
A0A212FNQ0
A0A194QVZ7
A0A194QLA7
+ More
A0A2A4K7Z3 A0A212EYQ0 A0A1Y1MD44 A0A1Y1M948 A0A034WK37 A0A0K8UP59 W8BJ09 A0A0R3NW20 A0A0J7L0V3 A0A1B6JFE9 A0A154PM38 A0A182F546 A0A2M3ZC89 A0A2M3ZCE1 A0A1I8PD91 B4KHR2 B4JQ02 F7BTW4 A0A139WEE8 B4LVB9 F6YPW6 A0A087ZTP2 A0A0L7RBC8 E2B927 Q7Q128 K7FL94 A0A0C9S2T5 K7FLD6 A0A195D6Q8 B4Q9A9 B4HWQ2 Q29R29 Q9VKV3 A0A151I0E7 E1JHE3 A0A0N7ZCV3 A0A0P4WFN0 B4P0P5 A0A310SHM2 A0A3B3QJJ2 B4MUB3 Q8MSF8 A0A091PJB2 A0A3B3QKE1 A0A158NJN3 A0A182PW42 A0A3L8DX48 B3N5F4 D3ZD40 A0A026W198 A0A386H774 A0A3L8SXQ7 H0ZLV6 A0A195FPF2 A0A3Q3KQK2 L8Y4A4 A0A091MGK3 G3SMX7 A0A3B3CPB9 H0X0C8 A0A093DQZ5 A0A182JER1 A0A1U7QTI5 A0A091TDY8 A0A1U7QQV9 A0A3B3CP76 Q9EPX2 B7ZN28 A0A093IU08 A0A3B3DZ05 A0A340XID0 A0A2J8PJB5 A0A2R9CCA8 A0A2R9CCH0 H2R995 A0A2I3SKU7 A0A2J8T4F8 A0A2J8T4D1 H2NLQ3 A0A2J8T4D6 A0A195EKD7 A0A2I3T2D6 A0A2J8PJD7 A0A096NVY9 A0A2K5NMV5 A0A2J8PJD5 A0A2K5NN26 A0A2J8T4C5 A0A2K5NMV9 A0A2K6TCZ1 A0A2K6TCW2 A0A1B6M8Z1
A0A2A4K7Z3 A0A212EYQ0 A0A1Y1MD44 A0A1Y1M948 A0A034WK37 A0A0K8UP59 W8BJ09 A0A0R3NW20 A0A0J7L0V3 A0A1B6JFE9 A0A154PM38 A0A182F546 A0A2M3ZC89 A0A2M3ZCE1 A0A1I8PD91 B4KHR2 B4JQ02 F7BTW4 A0A139WEE8 B4LVB9 F6YPW6 A0A087ZTP2 A0A0L7RBC8 E2B927 Q7Q128 K7FL94 A0A0C9S2T5 K7FLD6 A0A195D6Q8 B4Q9A9 B4HWQ2 Q29R29 Q9VKV3 A0A151I0E7 E1JHE3 A0A0N7ZCV3 A0A0P4WFN0 B4P0P5 A0A310SHM2 A0A3B3QJJ2 B4MUB3 Q8MSF8 A0A091PJB2 A0A3B3QKE1 A0A158NJN3 A0A182PW42 A0A3L8DX48 B3N5F4 D3ZD40 A0A026W198 A0A386H774 A0A3L8SXQ7 H0ZLV6 A0A195FPF2 A0A3Q3KQK2 L8Y4A4 A0A091MGK3 G3SMX7 A0A3B3CPB9 H0X0C8 A0A093DQZ5 A0A182JER1 A0A1U7QTI5 A0A091TDY8 A0A1U7QQV9 A0A3B3CP76 Q9EPX2 B7ZN28 A0A093IU08 A0A3B3DZ05 A0A340XID0 A0A2J8PJB5 A0A2R9CCA8 A0A2R9CCH0 H2R995 A0A2I3SKU7 A0A2J8T4F8 A0A2J8T4D1 H2NLQ3 A0A2J8T4D6 A0A195EKD7 A0A2I3T2D6 A0A2J8PJD7 A0A096NVY9 A0A2K5NMV5 A0A2J8PJD5 A0A2K5NN26 A0A2J8T4C5 A0A2K5NMV9 A0A2K6TCZ1 A0A2K6TCW2 A0A1B6M8Z1
PDB
3GHN
E-value=1.51013e-12,
Score=172
Ontologies
Topology
Subcellular location
Secreted
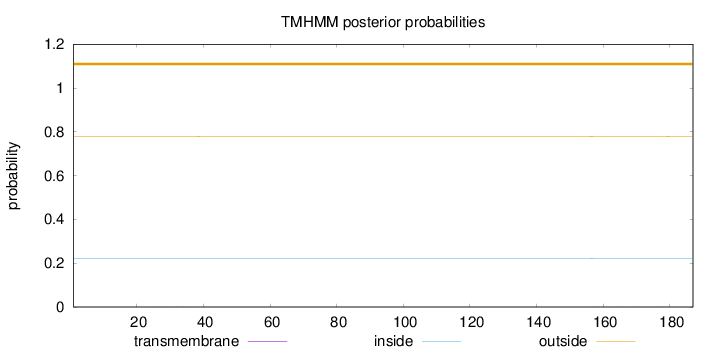
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00918000000000001
Exp number, first 60 AAs:
0.00298
Total prob of N-in:
0.22201
outside
1 - 187
Population Genetic Test Statistics
Pi
225.759971
Theta
171.39937
Tajima's D
1.108638
CLR
0.314012
CSRT
0.691365431728414
Interpretation
Uncertain
