Gene
KWMTBOMO11486 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001915
Annotation
proteasome_subunit_alpha_type-4_[Papilio_xuthus]
Full name
Proteasome subunit alpha type
+ More
Proteasome endopeptidase complex
Proteasome subunit beta
Proteasome endopeptidase complex
Proteasome subunit beta
Location in the cell
Cytoplasmic Reliability : 1.964
Sequence
CDS
ATGGCACGACGTTATGATACCCGTACAACCATATTCTCGCCGGAAGGTCGTCTGTATCAAGTCGAATATGCTATGGAAGCTATAAGTCATGCGGGAACATCGCTGGGTATTTTAGCAACGGATGGTATTCTTTTGGCTGCAGAACGCAGGAACACCAATAAACTTCTAGATGAAGTTTTCTTTTCTGAAAAAATCTACAAGCTCAATGATGACATGGTCTGTTCAGTCGCTGGTATAACATCGGATGCTAACGTTTTGACAAACGAATTAAGACTGATCGCTCAAAGATACCTGCTGCAATATGGCGAATCTATCCCTTGTGAGCAATTAGTCTCCTGGCTTTGTGATGTGAAACAAGCTTACACACAATATGGAGGTAAAAGGCCCTTTGGAGTCTCAATTCTGTATATGGGCTGGGATAAGCACTATGGTTATCAGCTCTACCAGTCTGATCCCAGTGGAAACTATGGGGGGTGGAAAGCAACATGCATCGGTAACAATAGTGCTGCAGCAGTTTCAAGTTTAAAACAGGAGTACAAGGAAAATGAGACAACTCTGGCAGAAGCACAGGCCCTGGCAATCAAAGTACTTAGCAAGACTCTGGATATGACTAAACTGACTCCAGAAAAAGTCGAAATGGCAACATTAACTCGTAAGGACAACAAGACTATCATCAGGATTCTAACGAATACAGAAGTTGAGAAACTCATTGCCGAATTCGAAAAGACCGAAGCTGAAGCTGAAGCTGCCAAGAAACAGCCTGCTAAATCTTAA
Protein
MARRYDTRTTIFSPEGRLYQVEYAMEAISHAGTSLGILATDGILLAAERRNTNKLLDEVFFSEKIYKLNDDMVCSVAGITSDANVLTNELRLIAQRYLLQYGESIPCEQLVSWLCDVKQAYTQYGGKRPFGVSILYMGWDKHYGYQLYQSDPSGNYGGWKATCIGNNSAAAVSSLKQEYKENETTLAEAQALAIKVLSKTLDMTKLTPEKVEMATLTRKDNKTIIRILTNTEVEKLIAEFEKTEAEAEAAKKQPAKS
Summary
Description
The proteasome is a multicatalytic proteinase complex which is characterized by its ability to cleave peptides with Arg, Phe, Tyr, Leu, and Glu adjacent to the leaving group at neutral or slightly basic pH.
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Subunit
The 20S proteasome core is composed of 28 subunits that are arranged in four stacked rings, resulting in a barrel-shaped structure. The two end rings are each formed by seven alpha subunits, and the two central rings are each formed by seven beta subunits.
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
Similarity
Belongs to the peptidase T1A family.
Belongs to the peptidase T1B family.
Belongs to the peptidase T1B family.
Uniprot
I4DK20
A0A3G1T1M6
A0A3S2NIA4
A0A2A4K7Z4
A0A212F5Q4
A0A2H1WB52
+ More
Q60FS3 A0A1Y1MKT5 A0A0C9QWS6 A0A2J7Q0B4 A0A1W4WBV2 D6WSJ0 V5GN54 A0A1B6DW66 A0A026WIN6 K7J976 A0A1B6JBP7 A0A1B6GUW9 A0A023GH28 G3MLT2 E9IBT3 A0A194QXM5 A0A2P6LEC0 A0A087SWI0 A0A023FY32 E0VLL3 A0A1E1X2Z1 S4PTA8 E2A3M2 F4W8N1 J3JYC7 B0WVH1 A0A023FGZ7 Q16TY4 A0A0K8RM20 A0A195EHJ1 A0A131XL10 A0A224YS45 A0A131Z3Y6 L7M2B5 V9IIY5 T1DGS8 A0A1Q3FFJ4 A0A023ENX8 A0A194QK15 A0A2M4ANC0 A0A2M4BX52 A0A2M4AAR7 A0A2M3ZHB4 A0A2A3E1A3 E9HJU6 A0A182YR47 A0A2C9H4D1 A0A182FB12 A0A2K6VB16 A0A1L8DSU3 A0A240SY42 A0A1Q3FFJ7 A0A1Z5KXF2 A0A182KFM1 A0A182PTW1 A0A158NKS7 A0A182WM26 A0A182XEH6 A0A182VKR5 A0A182KNN7 A0A182HQL9 A0A182ULJ9 Q7PPI7 A0A182LS11 A0A182RE57 A0A2M4CTT7 A0A182J2X4 A0A336KJN3 A0A2M4AAK3 A0A182N1Y2 A0A2R5LIV4 A0A182QKR3 A0A0J7KT49 A0A3G5APG7 A0A0N8CTG7 A0A0N8DRX8 A0A0P6HIT7 A0A0K8TSN2 A0A0P5PBU4 A0A0P5ASC4 U5EWD4 A0A293LMM4 A0A088A859 A0A3G5AP11 T1K2I7 A0A1V9XRM7 A0A2G8LMW8 A0A0P6I7G1 H3CW10 A0A3Q3M748 A0A3P9H0Q5 H2L7M2 A0A3P9JWC0 A0A3S2Q941
Q60FS3 A0A1Y1MKT5 A0A0C9QWS6 A0A2J7Q0B4 A0A1W4WBV2 D6WSJ0 V5GN54 A0A1B6DW66 A0A026WIN6 K7J976 A0A1B6JBP7 A0A1B6GUW9 A0A023GH28 G3MLT2 E9IBT3 A0A194QXM5 A0A2P6LEC0 A0A087SWI0 A0A023FY32 E0VLL3 A0A1E1X2Z1 S4PTA8 E2A3M2 F4W8N1 J3JYC7 B0WVH1 A0A023FGZ7 Q16TY4 A0A0K8RM20 A0A195EHJ1 A0A131XL10 A0A224YS45 A0A131Z3Y6 L7M2B5 V9IIY5 T1DGS8 A0A1Q3FFJ4 A0A023ENX8 A0A194QK15 A0A2M4ANC0 A0A2M4BX52 A0A2M4AAR7 A0A2M3ZHB4 A0A2A3E1A3 E9HJU6 A0A182YR47 A0A2C9H4D1 A0A182FB12 A0A2K6VB16 A0A1L8DSU3 A0A240SY42 A0A1Q3FFJ7 A0A1Z5KXF2 A0A182KFM1 A0A182PTW1 A0A158NKS7 A0A182WM26 A0A182XEH6 A0A182VKR5 A0A182KNN7 A0A182HQL9 A0A182ULJ9 Q7PPI7 A0A182LS11 A0A182RE57 A0A2M4CTT7 A0A182J2X4 A0A336KJN3 A0A2M4AAK3 A0A182N1Y2 A0A2R5LIV4 A0A182QKR3 A0A0J7KT49 A0A3G5APG7 A0A0N8CTG7 A0A0N8DRX8 A0A0P6HIT7 A0A0K8TSN2 A0A0P5PBU4 A0A0P5ASC4 U5EWD4 A0A293LMM4 A0A088A859 A0A3G5AP11 T1K2I7 A0A1V9XRM7 A0A2G8LMW8 A0A0P6I7G1 H3CW10 A0A3Q3M748 A0A3P9H0Q5 H2L7M2 A0A3P9JWC0 A0A3S2Q941
EC Number
3.4.25.1
Pubmed
22651552
22118469
28004739
18362917
19820115
24508170
+ More
30249741 20075255 22216098 21282665 26354079 20566863 28503490 23622113 20798317 21719571 22516182 23537049 17510324 28049606 28797301 26830274 25576852 24330624 24945155 26483478 21292972 25244985 20920257 28528879 21347285 20966253 12364791 14747013 17210077 26369729 28327890 29023486 15496914 17554307
30249741 20075255 22216098 21282665 26354079 20566863 28503490 23622113 20798317 21719571 22516182 23537049 17510324 28049606 28797301 26830274 25576852 24330624 24945155 26483478 21292972 25244985 20920257 28528879 21347285 20966253 12364791 14747013 17210077 26369729 28327890 29023486 15496914 17554307
EMBL
AK401638
BAM18260.1
MG992425
AXY94863.1
RSAL01000086
RVE48288.1
+ More
NWSH01000085 PCG79782.1 AGBW02010153 OWR49066.1 ODYU01007425 SOQ50186.1 AB189026 BAD52258.1 GEZM01028378 JAV86333.1 GBYB01005127 JAG74894.1 NEVH01019983 PNF22021.1 KQ971354 EFA05912.1 GALX01005474 JAB62992.1 GEDC01007419 JAS29879.1 KK107183 QOIP01000011 EZA55885.1 RLU17216.1 AAZX01011560 GECU01011253 JAS96453.1 GECZ01003562 JAS66207.1 GBBM01002141 JAC33277.1 JO842833 AEO34450.1 GL762137 EFZ21978.1 KQ461073 KPJ09710.1 MWRG01000153 PRD36921.1 KK112265 KFM57219.1 GBBL01001682 JAC25638.1 DS235274 EEB14269.1 GFAC01005782 JAT93406.1 GAIX01012363 JAA80197.1 GL436457 EFN71914.1 GL887908 EGI69522.1 APGK01028660 BT128253 KB740694 KB632194 AEE63213.1 ENN79476.1 ERL89890.1 DS232126 EDS35577.1 GBBK01003311 JAC21171.1 CH477637 EAT37982.1 GADI01002219 JAA71589.1 KQ978881 KYN27738.1 GEFH01002215 JAP66366.1 GFPF01005496 MAA16642.1 GEDV01002438 JAP86119.1 GACK01007027 JAA58007.1 JR047620 AEY60461.1 GALA01000077 JAA94775.1 GFDL01008731 JAV26314.1 JXUM01024338 GAPW01003439 KQ560687 JAC10159.1 KXJ81276.1 KQ458575 KPJ05719.1 GGFK01008962 MBW42283.1 GGFJ01008519 MBW57660.1 GGFK01004519 MBW37840.1 GGFM01007195 MBW27946.1 KZ288473 PBC25304.1 GL732664 EFX68008.1 GGFL01004417 MBW68595.1 GFDF01004637 JAV09447.1 AJWK01030742 GFDL01008730 JAV26315.1 GFJQ02007147 JAV99822.1 ADTU01018781 APCN01004657 AAAB01008948 EAA10351.5 AXCM01003001 GGFL01004413 MBW68591.1 UFQS01000509 UFQT01000509 SSX04491.1 SSX24855.1 GGFK01004449 MBW37770.1 GGLE01005131 MBY09257.1 AXCN02000977 LBMM01003526 KMQ93419.1 MH979785 AYV89240.1 GDIP01100511 LRGB01000930 JAM03204.1 KZS15037.1 GDIP01006823 JAM96892.1 GDIQ01042133 JAN52604.1 GDAI01000470 JAI17133.1 GDIQ01152492 JAK99233.1 GDIP01194751 JAJ28651.1 GANO01001465 JAB58406.1 GFWV01003225 MAA27955.1 MH990543 AYV89090.1 CAEY01001363 MNPL01005442 OQR76018.1 MRZV01000028 PIK61603.1 GDIQ01008583 JAN86154.1 CM012439 RVE74211.1
NWSH01000085 PCG79782.1 AGBW02010153 OWR49066.1 ODYU01007425 SOQ50186.1 AB189026 BAD52258.1 GEZM01028378 JAV86333.1 GBYB01005127 JAG74894.1 NEVH01019983 PNF22021.1 KQ971354 EFA05912.1 GALX01005474 JAB62992.1 GEDC01007419 JAS29879.1 KK107183 QOIP01000011 EZA55885.1 RLU17216.1 AAZX01011560 GECU01011253 JAS96453.1 GECZ01003562 JAS66207.1 GBBM01002141 JAC33277.1 JO842833 AEO34450.1 GL762137 EFZ21978.1 KQ461073 KPJ09710.1 MWRG01000153 PRD36921.1 KK112265 KFM57219.1 GBBL01001682 JAC25638.1 DS235274 EEB14269.1 GFAC01005782 JAT93406.1 GAIX01012363 JAA80197.1 GL436457 EFN71914.1 GL887908 EGI69522.1 APGK01028660 BT128253 KB740694 KB632194 AEE63213.1 ENN79476.1 ERL89890.1 DS232126 EDS35577.1 GBBK01003311 JAC21171.1 CH477637 EAT37982.1 GADI01002219 JAA71589.1 KQ978881 KYN27738.1 GEFH01002215 JAP66366.1 GFPF01005496 MAA16642.1 GEDV01002438 JAP86119.1 GACK01007027 JAA58007.1 JR047620 AEY60461.1 GALA01000077 JAA94775.1 GFDL01008731 JAV26314.1 JXUM01024338 GAPW01003439 KQ560687 JAC10159.1 KXJ81276.1 KQ458575 KPJ05719.1 GGFK01008962 MBW42283.1 GGFJ01008519 MBW57660.1 GGFK01004519 MBW37840.1 GGFM01007195 MBW27946.1 KZ288473 PBC25304.1 GL732664 EFX68008.1 GGFL01004417 MBW68595.1 GFDF01004637 JAV09447.1 AJWK01030742 GFDL01008730 JAV26315.1 GFJQ02007147 JAV99822.1 ADTU01018781 APCN01004657 AAAB01008948 EAA10351.5 AXCM01003001 GGFL01004413 MBW68591.1 UFQS01000509 UFQT01000509 SSX04491.1 SSX24855.1 GGFK01004449 MBW37770.1 GGLE01005131 MBY09257.1 AXCN02000977 LBMM01003526 KMQ93419.1 MH979785 AYV89240.1 GDIP01100511 LRGB01000930 JAM03204.1 KZS15037.1 GDIP01006823 JAM96892.1 GDIQ01042133 JAN52604.1 GDAI01000470 JAI17133.1 GDIQ01152492 JAK99233.1 GDIP01194751 JAJ28651.1 GANO01001465 JAB58406.1 GFWV01003225 MAA27955.1 MH990543 AYV89090.1 CAEY01001363 MNPL01005442 OQR76018.1 MRZV01000028 PIK61603.1 GDIQ01008583 JAN86154.1 CM012439 RVE74211.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000235965
UP000192223
UP000007266
+ More
UP000053097 UP000279307 UP000002358 UP000053240 UP000054359 UP000009046 UP000000311 UP000007755 UP000019118 UP000030742 UP000002320 UP000008820 UP000078492 UP000069940 UP000249989 UP000053268 UP000242457 UP000000305 UP000076408 UP000075901 UP000069272 UP000092461 UP000075881 UP000075885 UP000005205 UP000075920 UP000076407 UP000075903 UP000075882 UP000075840 UP000075902 UP000007062 UP000075883 UP000075900 UP000075880 UP000075884 UP000075886 UP000036403 UP000076858 UP000005203 UP000015104 UP000192247 UP000230750 UP000007303 UP000261640 UP000265200 UP000001038 UP000265180
UP000053097 UP000279307 UP000002358 UP000053240 UP000054359 UP000009046 UP000000311 UP000007755 UP000019118 UP000030742 UP000002320 UP000008820 UP000078492 UP000069940 UP000249989 UP000053268 UP000242457 UP000000305 UP000076408 UP000075901 UP000069272 UP000092461 UP000075881 UP000075885 UP000005205 UP000075920 UP000076407 UP000075903 UP000075882 UP000075840 UP000075902 UP000007062 UP000075883 UP000075900 UP000075880 UP000075884 UP000075886 UP000036403 UP000076858 UP000005203 UP000015104 UP000192247 UP000230750 UP000007303 UP000261640 UP000265200 UP000001038 UP000265180
PRIDE
Interpro
Gene 3D
ProteinModelPortal
I4DK20
A0A3G1T1M6
A0A3S2NIA4
A0A2A4K7Z4
A0A212F5Q4
A0A2H1WB52
+ More
Q60FS3 A0A1Y1MKT5 A0A0C9QWS6 A0A2J7Q0B4 A0A1W4WBV2 D6WSJ0 V5GN54 A0A1B6DW66 A0A026WIN6 K7J976 A0A1B6JBP7 A0A1B6GUW9 A0A023GH28 G3MLT2 E9IBT3 A0A194QXM5 A0A2P6LEC0 A0A087SWI0 A0A023FY32 E0VLL3 A0A1E1X2Z1 S4PTA8 E2A3M2 F4W8N1 J3JYC7 B0WVH1 A0A023FGZ7 Q16TY4 A0A0K8RM20 A0A195EHJ1 A0A131XL10 A0A224YS45 A0A131Z3Y6 L7M2B5 V9IIY5 T1DGS8 A0A1Q3FFJ4 A0A023ENX8 A0A194QK15 A0A2M4ANC0 A0A2M4BX52 A0A2M4AAR7 A0A2M3ZHB4 A0A2A3E1A3 E9HJU6 A0A182YR47 A0A2C9H4D1 A0A182FB12 A0A2K6VB16 A0A1L8DSU3 A0A240SY42 A0A1Q3FFJ7 A0A1Z5KXF2 A0A182KFM1 A0A182PTW1 A0A158NKS7 A0A182WM26 A0A182XEH6 A0A182VKR5 A0A182KNN7 A0A182HQL9 A0A182ULJ9 Q7PPI7 A0A182LS11 A0A182RE57 A0A2M4CTT7 A0A182J2X4 A0A336KJN3 A0A2M4AAK3 A0A182N1Y2 A0A2R5LIV4 A0A182QKR3 A0A0J7KT49 A0A3G5APG7 A0A0N8CTG7 A0A0N8DRX8 A0A0P6HIT7 A0A0K8TSN2 A0A0P5PBU4 A0A0P5ASC4 U5EWD4 A0A293LMM4 A0A088A859 A0A3G5AP11 T1K2I7 A0A1V9XRM7 A0A2G8LMW8 A0A0P6I7G1 H3CW10 A0A3Q3M748 A0A3P9H0Q5 H2L7M2 A0A3P9JWC0 A0A3S2Q941
Q60FS3 A0A1Y1MKT5 A0A0C9QWS6 A0A2J7Q0B4 A0A1W4WBV2 D6WSJ0 V5GN54 A0A1B6DW66 A0A026WIN6 K7J976 A0A1B6JBP7 A0A1B6GUW9 A0A023GH28 G3MLT2 E9IBT3 A0A194QXM5 A0A2P6LEC0 A0A087SWI0 A0A023FY32 E0VLL3 A0A1E1X2Z1 S4PTA8 E2A3M2 F4W8N1 J3JYC7 B0WVH1 A0A023FGZ7 Q16TY4 A0A0K8RM20 A0A195EHJ1 A0A131XL10 A0A224YS45 A0A131Z3Y6 L7M2B5 V9IIY5 T1DGS8 A0A1Q3FFJ4 A0A023ENX8 A0A194QK15 A0A2M4ANC0 A0A2M4BX52 A0A2M4AAR7 A0A2M3ZHB4 A0A2A3E1A3 E9HJU6 A0A182YR47 A0A2C9H4D1 A0A182FB12 A0A2K6VB16 A0A1L8DSU3 A0A240SY42 A0A1Q3FFJ7 A0A1Z5KXF2 A0A182KFM1 A0A182PTW1 A0A158NKS7 A0A182WM26 A0A182XEH6 A0A182VKR5 A0A182KNN7 A0A182HQL9 A0A182ULJ9 Q7PPI7 A0A182LS11 A0A182RE57 A0A2M4CTT7 A0A182J2X4 A0A336KJN3 A0A2M4AAK3 A0A182N1Y2 A0A2R5LIV4 A0A182QKR3 A0A0J7KT49 A0A3G5APG7 A0A0N8CTG7 A0A0N8DRX8 A0A0P6HIT7 A0A0K8TSN2 A0A0P5PBU4 A0A0P5ASC4 U5EWD4 A0A293LMM4 A0A088A859 A0A3G5AP11 T1K2I7 A0A1V9XRM7 A0A2G8LMW8 A0A0P6I7G1 H3CW10 A0A3Q3M748 A0A3P9H0Q5 H2L7M2 A0A3P9JWC0 A0A3S2Q941
PDB
6AVO
E-value=2.33316e-112,
Score=1035
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
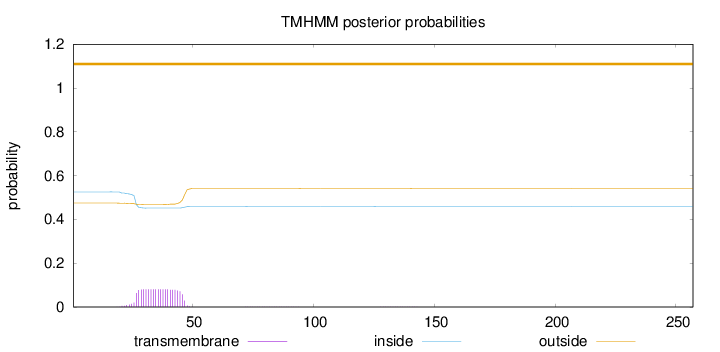
Length:
257
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.66481
Exp number, first 60 AAs:
1.65302
Total prob of N-in:
0.52558
outside
1 - 257
Population Genetic Test Statistics
Pi
202.446583
Theta
187.216036
Tajima's D
0.262753
CLR
0.419223
CSRT
0.445077746112694
Interpretation
Uncertain
