Pre Gene Modal
BGIBMGA001916
Annotation
embryonic_development_factor_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.482 Nuclear Reliability : 1.914
Sequence
CDS
ATGGCAATAGAGAAACCCAAATTGTCCACAGTTCCTGTACCAAAGAAATCTCGCTTAGAAATGAATGTCAGACAGCTCCTAAGTAAATGTGAACTTATAGCCAAAGAGGAATCAATAGACGGGAACTGGCGTCTAAAAAAATATGTGGAATCTTTAAGCAGTATGATTACGGAACTAAAAACTGGACCTGATAAACCATCAAAAGATATACTGTCTGAGTATTCAAAAAGAGCGGCTTTCTTAAAAGGTGTTGTTCAAACTGCTACACTGAGCACTCCTATTGAGAAGCTTGAAGCAGTTCAACAACTATCGCACGGTGCAGCTACAATGTCAGTTGATGCAACACAAGAAATACATCAAAAGACTGTTGCTAAGTATGGAGTAGAATTAAGATCTGAACTGTTTGGGTTGGATGATAATGATGACTCTCTCCGTAAAAGGAACATTATTAAAACACCAAATTTCACAAGCAACACTACCAGTCAGGAAGACATTGATTCAATTCTGAAATACCATCAGAACATGCAGGAGAAAGTCGCTGAGAACATGGTGATGCTTACAAAGAACTTGAAGGAGCAGTCACAGATTGCTAGTACAATTATTAAGGCAGATACAGAGGCATTAAAAAAATCATCAGACTTGACCGACCGTAACTTGTCATCACTGAAAGTGGAATCGGAGAGATTGCAGGAACACAGTCGAAGTGCCTGGAAATGTTGGCTCTGGATCATGTTAGCTGTGGTCATGGTTATATTTATTAATATGGTCCTGTTTATGAAGGTTATGAAGAAAAGAAAATACGAACTTTAA
Protein
MAIEKPKLSTVPVPKKSRLEMNVRQLLSKCELIAKEESIDGNWRLKKYVESLSSMITELKTGPDKPSKDILSEYSKRAAFLKGVVQTATLSTPIEKLEAVQQLSHGAATMSVDATQEIHQKTVAKYGVELRSELFGLDDNDDSLRKRNIIKTPNFTSNTTSQEDIDSILKYHQNMQEKVAENMVMLTKNLKEQSQIASTIIKADTEALKKSSDLTDRNLSSLKVESERLQEHSRSAWKCWLWIMLAVVMVIFINMVLFMKVMKKRKYEL
Summary
Uniprot
Q1HQ91
H9IXD3
A0A1E1WAR8
A0A212F5Q0
S4PSC1
A0A194R1I4
+ More
A0A2H1WAX8 A0A194QJH1 A0A067QMR0 A0A1B6K5L5 A0A1B6FEZ9 A0A2J7PMC2 A0A195BXY4 A0A151JSG9 A0A158NL54 A0A195BG00 A0A195F117 F4WFW2 A0A2A3EPC9 A0A0L7R7H9 A0A154PH91 A0A224XVG0 A0A069DQK0 E2B6I7 A0A151XBG6 A0A023EZG4 A0A3L8D8V2 A0A232F4G7 E2AHZ8 A0A026WSG2 A0A0N7Z8V4 A0A310SCL0 A0A1W7R842 A0A1S4FGU0 Q171U4 A0A0M8ZW64 W8CE16 E0VMG0 A0A1D2NH08 U5EV67 A0A1Q3FBW1 A0A0K8USX1 A0A034WIZ9 B0VZ75 A0A2U3W8P9 A0A2Y9IYP9 M3YWW7 M3VVT3 A0A0C9RE41 A0A3Q7X0V3 A0A0A1WX35 A0A2F0B3Y3 A0A384B784 A0A1B6D1L4 A0A2Y9RJH1 A0A2U3W8Q7 A0A0C9RY72 L8HQE4 Q148D2 F6QDZ3 G1PPW4 A0A2U3Z1C5 A0A2Y9F9L9 U3E9N1 A0A2Y9GLI1 A0A3Q7Q3J1 A0A2Y9LXF0 W5Q8P3 A0A2K6E895 F6PM56 F6PM29 G1LUY3 A0A087TJK2 A0A2K5D2C3 A0A341D8G0 A0A2K5X308 A0A0D9R025 A0A0A0MWZ9 H9Z3S9 A0A2K6G808 A0A2K6QBP3 A0A2K6RZ02 G9KWQ8 L9KXH3 G3R1W2 G1R106 A0A2R9A2D6 A0A2K5KDV0 K7CL56 B0BNG6 A0A2K5Q4L4 F1LQ22 A0A2K5MPZ2 A0A3Q7RTQ0 A0A287D873 A0A250YE68 F6X511 A0A3M6TJV5 H2NXZ6 A0A1I8N663
A0A2H1WAX8 A0A194QJH1 A0A067QMR0 A0A1B6K5L5 A0A1B6FEZ9 A0A2J7PMC2 A0A195BXY4 A0A151JSG9 A0A158NL54 A0A195BG00 A0A195F117 F4WFW2 A0A2A3EPC9 A0A0L7R7H9 A0A154PH91 A0A224XVG0 A0A069DQK0 E2B6I7 A0A151XBG6 A0A023EZG4 A0A3L8D8V2 A0A232F4G7 E2AHZ8 A0A026WSG2 A0A0N7Z8V4 A0A310SCL0 A0A1W7R842 A0A1S4FGU0 Q171U4 A0A0M8ZW64 W8CE16 E0VMG0 A0A1D2NH08 U5EV67 A0A1Q3FBW1 A0A0K8USX1 A0A034WIZ9 B0VZ75 A0A2U3W8P9 A0A2Y9IYP9 M3YWW7 M3VVT3 A0A0C9RE41 A0A3Q7X0V3 A0A0A1WX35 A0A2F0B3Y3 A0A384B784 A0A1B6D1L4 A0A2Y9RJH1 A0A2U3W8Q7 A0A0C9RY72 L8HQE4 Q148D2 F6QDZ3 G1PPW4 A0A2U3Z1C5 A0A2Y9F9L9 U3E9N1 A0A2Y9GLI1 A0A3Q7Q3J1 A0A2Y9LXF0 W5Q8P3 A0A2K6E895 F6PM56 F6PM29 G1LUY3 A0A087TJK2 A0A2K5D2C3 A0A341D8G0 A0A2K5X308 A0A0D9R025 A0A0A0MWZ9 H9Z3S9 A0A2K6G808 A0A2K6QBP3 A0A2K6RZ02 G9KWQ8 L9KXH3 G3R1W2 G1R106 A0A2R9A2D6 A0A2K5KDV0 K7CL56 B0BNG6 A0A2K5Q4L4 F1LQ22 A0A2K5MPZ2 A0A3Q7RTQ0 A0A287D873 A0A250YE68 F6X511 A0A3M6TJV5 H2NXZ6 A0A1I8N663
Pubmed
19121390
22118469
23622113
26354079
24845553
21347285
+ More
21719571 26334808 20798317 25474469 30249741 28648823 24508170 27129103 17510324 24495485 20566863 27289101 25348373 17975172 25830018 22751099 19393038 21993624 25243066 20809919 17431167 25319552 20010809 25362486 23236062 23385571 22398555 22722832 15489334 15057822 28087693 16341006 30382153 25315136
21719571 26334808 20798317 25474469 30249741 28648823 24508170 27129103 17510324 24495485 20566863 27289101 25348373 17975172 25830018 22751099 19393038 21993624 25243066 20809919 17431167 25319552 20010809 25362486 23236062 23385571 22398555 22722832 15489334 15057822 28087693 16341006 30382153 25315136
EMBL
DQ443161
ABF51250.1
BABH01017977
GDQN01006954
JAT84100.1
AGBW02010153
+ More
OWR49065.1 GAIX01000065 JAA92495.1 KQ461073 KPJ09711.1 ODYU01007425 SOQ50187.1 KQ458575 KPJ05718.1 KK853141 KDR10759.1 GECU01000969 JAT06738.1 GECZ01021002 JAS48767.1 NEVH01024189 PNF17476.1 KQ978501 KYM93519.1 KQ978530 KYN30313.1 ADTU01019257 KQ976488 KYM83517.1 KQ981866 KYN34076.1 GL888128 EGI66729.1 KZ288209 PBC32989.1 KQ414642 KOC66716.1 KQ434904 KZC11182.1 GFTR01004006 JAW12420.1 GBGD01002704 JAC86185.1 GL445975 EFN88679.1 KQ982320 KYQ57726.1 GBBI01004084 JAC14628.1 QOIP01000011 RLU16552.1 NNAY01001043 OXU25370.1 GL439621 EFN66906.1 KK107111 EZA58913.1 GDKW01002299 JAI54296.1 KQ764051 OAD54618.1 GEHC01000321 JAV47324.1 CH477446 EAT40763.1 KQ435843 KOX71286.1 GAMC01000981 JAC05575.1 DS235307 EEB14566.1 LJIJ01000041 ODN04541.1 GANO01001158 JAB58713.1 GFDL01009981 JAV25064.1 GDHF01022859 JAI29455.1 GAKP01005224 JAC53728.1 DS231813 EDS25720.1 AEYP01051466 AANG04000016 GBYB01005226 JAG74993.1 GBXI01011314 JAD02978.1 NTJE010065715 MBV97730.1 GEDC01017728 JAS19570.1 GBYB01014215 JAG83982.1 JH883666 ELR46083.1 BC118451 AAI18452.1 AAPE02039128 GAMT01001669 GAMS01009743 GAMQ01000594 JAB10192.1 JAB13393.1 JAB41257.1 AMGL01090895 JSUE03021071 JU321884 AFE65640.1 ACTA01056567 ACTA01064567 KK115505 KFM65291.1 AQIA01034692 AQIB01141366 AHZZ02013753 JU473791 JV044433 AFH30595.1 AFI34504.1 JP020739 AES09337.1 KB320623 ELW67189.1 CABD030112158 CABD030112159 CABD030112160 ADFV01090047 ADFV01090048 ADFV01090049 AJFE02060627 GABC01010107 GABF01004951 GABD01010765 GABE01011581 NBAG03000278 JAA01231.1 JAA17194.1 JAA22335.1 JAA33158.1 PNI50698.1 PNI50699.1 BC158812 AAI58813.1 AC125838 AGTP01071372 GFFW01002871 JAV41917.1 AAEX03012299 RCHS01003477 RMX41558.1 ABGA01183387 ABGA01183388
OWR49065.1 GAIX01000065 JAA92495.1 KQ461073 KPJ09711.1 ODYU01007425 SOQ50187.1 KQ458575 KPJ05718.1 KK853141 KDR10759.1 GECU01000969 JAT06738.1 GECZ01021002 JAS48767.1 NEVH01024189 PNF17476.1 KQ978501 KYM93519.1 KQ978530 KYN30313.1 ADTU01019257 KQ976488 KYM83517.1 KQ981866 KYN34076.1 GL888128 EGI66729.1 KZ288209 PBC32989.1 KQ414642 KOC66716.1 KQ434904 KZC11182.1 GFTR01004006 JAW12420.1 GBGD01002704 JAC86185.1 GL445975 EFN88679.1 KQ982320 KYQ57726.1 GBBI01004084 JAC14628.1 QOIP01000011 RLU16552.1 NNAY01001043 OXU25370.1 GL439621 EFN66906.1 KK107111 EZA58913.1 GDKW01002299 JAI54296.1 KQ764051 OAD54618.1 GEHC01000321 JAV47324.1 CH477446 EAT40763.1 KQ435843 KOX71286.1 GAMC01000981 JAC05575.1 DS235307 EEB14566.1 LJIJ01000041 ODN04541.1 GANO01001158 JAB58713.1 GFDL01009981 JAV25064.1 GDHF01022859 JAI29455.1 GAKP01005224 JAC53728.1 DS231813 EDS25720.1 AEYP01051466 AANG04000016 GBYB01005226 JAG74993.1 GBXI01011314 JAD02978.1 NTJE010065715 MBV97730.1 GEDC01017728 JAS19570.1 GBYB01014215 JAG83982.1 JH883666 ELR46083.1 BC118451 AAI18452.1 AAPE02039128 GAMT01001669 GAMS01009743 GAMQ01000594 JAB10192.1 JAB13393.1 JAB41257.1 AMGL01090895 JSUE03021071 JU321884 AFE65640.1 ACTA01056567 ACTA01064567 KK115505 KFM65291.1 AQIA01034692 AQIB01141366 AHZZ02013753 JU473791 JV044433 AFH30595.1 AFI34504.1 JP020739 AES09337.1 KB320623 ELW67189.1 CABD030112158 CABD030112159 CABD030112160 ADFV01090047 ADFV01090048 ADFV01090049 AJFE02060627 GABC01010107 GABF01004951 GABD01010765 GABE01011581 NBAG03000278 JAA01231.1 JAA17194.1 JAA22335.1 JAA33158.1 PNI50698.1 PNI50699.1 BC158812 AAI58813.1 AC125838 AGTP01071372 GFFW01002871 JAV41917.1 AAEX03012299 RCHS01003477 RMX41558.1 ABGA01183387 ABGA01183388
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000027135
UP000235965
+ More
UP000078542 UP000078492 UP000005205 UP000078540 UP000078541 UP000007755 UP000242457 UP000053825 UP000076502 UP000008237 UP000075809 UP000279307 UP000215335 UP000000311 UP000053097 UP000008820 UP000053105 UP000009046 UP000094527 UP000002320 UP000245340 UP000248482 UP000000715 UP000011712 UP000286642 UP000261681 UP000248480 UP000009136 UP000001074 UP000245341 UP000248484 UP000008225 UP000248481 UP000286641 UP000248483 UP000002356 UP000233120 UP000006718 UP000008912 UP000054359 UP000233020 UP000252040 UP000233100 UP000029965 UP000028761 UP000233160 UP000233200 UP000233220 UP000011518 UP000001519 UP000001073 UP000240080 UP000233080 UP000233040 UP000002494 UP000233060 UP000286640 UP000005215 UP000002254 UP000275408 UP000001595 UP000095301
UP000078542 UP000078492 UP000005205 UP000078540 UP000078541 UP000007755 UP000242457 UP000053825 UP000076502 UP000008237 UP000075809 UP000279307 UP000215335 UP000000311 UP000053097 UP000008820 UP000053105 UP000009046 UP000094527 UP000002320 UP000245340 UP000248482 UP000000715 UP000011712 UP000286642 UP000261681 UP000248480 UP000009136 UP000001074 UP000245341 UP000248484 UP000008225 UP000248481 UP000286641 UP000248483 UP000002356 UP000233120 UP000006718 UP000008912 UP000054359 UP000233020 UP000252040 UP000233100 UP000029965 UP000028761 UP000233160 UP000233200 UP000233220 UP000011518 UP000001519 UP000001073 UP000240080 UP000233080 UP000233040 UP000002494 UP000233060 UP000286640 UP000005215 UP000002254 UP000275408 UP000001595 UP000095301
Interpro
Gene 3D
CDD
ProteinModelPortal
Q1HQ91
H9IXD3
A0A1E1WAR8
A0A212F5Q0
S4PSC1
A0A194R1I4
+ More
A0A2H1WAX8 A0A194QJH1 A0A067QMR0 A0A1B6K5L5 A0A1B6FEZ9 A0A2J7PMC2 A0A195BXY4 A0A151JSG9 A0A158NL54 A0A195BG00 A0A195F117 F4WFW2 A0A2A3EPC9 A0A0L7R7H9 A0A154PH91 A0A224XVG0 A0A069DQK0 E2B6I7 A0A151XBG6 A0A023EZG4 A0A3L8D8V2 A0A232F4G7 E2AHZ8 A0A026WSG2 A0A0N7Z8V4 A0A310SCL0 A0A1W7R842 A0A1S4FGU0 Q171U4 A0A0M8ZW64 W8CE16 E0VMG0 A0A1D2NH08 U5EV67 A0A1Q3FBW1 A0A0K8USX1 A0A034WIZ9 B0VZ75 A0A2U3W8P9 A0A2Y9IYP9 M3YWW7 M3VVT3 A0A0C9RE41 A0A3Q7X0V3 A0A0A1WX35 A0A2F0B3Y3 A0A384B784 A0A1B6D1L4 A0A2Y9RJH1 A0A2U3W8Q7 A0A0C9RY72 L8HQE4 Q148D2 F6QDZ3 G1PPW4 A0A2U3Z1C5 A0A2Y9F9L9 U3E9N1 A0A2Y9GLI1 A0A3Q7Q3J1 A0A2Y9LXF0 W5Q8P3 A0A2K6E895 F6PM56 F6PM29 G1LUY3 A0A087TJK2 A0A2K5D2C3 A0A341D8G0 A0A2K5X308 A0A0D9R025 A0A0A0MWZ9 H9Z3S9 A0A2K6G808 A0A2K6QBP3 A0A2K6RZ02 G9KWQ8 L9KXH3 G3R1W2 G1R106 A0A2R9A2D6 A0A2K5KDV0 K7CL56 B0BNG6 A0A2K5Q4L4 F1LQ22 A0A2K5MPZ2 A0A3Q7RTQ0 A0A287D873 A0A250YE68 F6X511 A0A3M6TJV5 H2NXZ6 A0A1I8N663
A0A2H1WAX8 A0A194QJH1 A0A067QMR0 A0A1B6K5L5 A0A1B6FEZ9 A0A2J7PMC2 A0A195BXY4 A0A151JSG9 A0A158NL54 A0A195BG00 A0A195F117 F4WFW2 A0A2A3EPC9 A0A0L7R7H9 A0A154PH91 A0A224XVG0 A0A069DQK0 E2B6I7 A0A151XBG6 A0A023EZG4 A0A3L8D8V2 A0A232F4G7 E2AHZ8 A0A026WSG2 A0A0N7Z8V4 A0A310SCL0 A0A1W7R842 A0A1S4FGU0 Q171U4 A0A0M8ZW64 W8CE16 E0VMG0 A0A1D2NH08 U5EV67 A0A1Q3FBW1 A0A0K8USX1 A0A034WIZ9 B0VZ75 A0A2U3W8P9 A0A2Y9IYP9 M3YWW7 M3VVT3 A0A0C9RE41 A0A3Q7X0V3 A0A0A1WX35 A0A2F0B3Y3 A0A384B784 A0A1B6D1L4 A0A2Y9RJH1 A0A2U3W8Q7 A0A0C9RY72 L8HQE4 Q148D2 F6QDZ3 G1PPW4 A0A2U3Z1C5 A0A2Y9F9L9 U3E9N1 A0A2Y9GLI1 A0A3Q7Q3J1 A0A2Y9LXF0 W5Q8P3 A0A2K6E895 F6PM56 F6PM29 G1LUY3 A0A087TJK2 A0A2K5D2C3 A0A341D8G0 A0A2K5X308 A0A0D9R025 A0A0A0MWZ9 H9Z3S9 A0A2K6G808 A0A2K6QBP3 A0A2K6RZ02 G9KWQ8 L9KXH3 G3R1W2 G1R106 A0A2R9A2D6 A0A2K5KDV0 K7CL56 B0BNG6 A0A2K5Q4L4 F1LQ22 A0A2K5MPZ2 A0A3Q7RTQ0 A0A287D873 A0A250YE68 F6X511 A0A3M6TJV5 H2NXZ6 A0A1I8N663
Ontologies
PANTHER
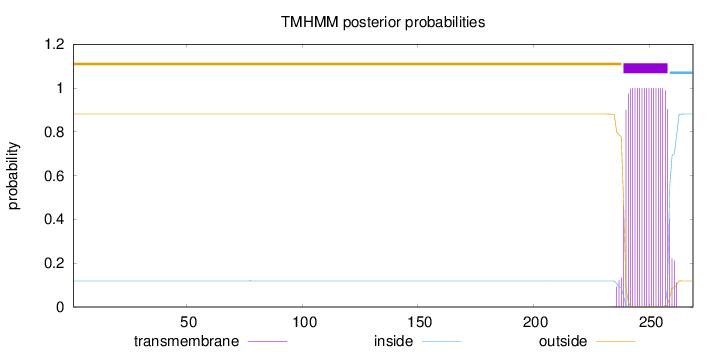
Topology
Length:
269
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.52709
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11909
outside
1 - 238
TMhelix
239 - 258
inside
259 - 269
Population Genetic Test Statistics
Pi
244.721616
Theta
179.99154
Tajima's D
1.205501
CLR
0
CSRT
0.713514324283786
Interpretation
Uncertain
