Gene
KWMTBOMO11484
Pre Gene Modal
BGIBMGA001905
Annotation
PREDICTED:_peptidyl-alpha-hydroxyglycine_alpha-amidating_lyase_2-like_[Papilio_xuthus]
Full name
Peptidyl-alpha-hydroxyglycine alpha-amidating lyase 2
Alternative Name
Peptidylamidoglycolate lyase 2
Location in the cell
Extracellular Reliability : 2.077
Sequence
CDS
ATGTTGTTCGTTTTACTGTACGCCGTAATTCTTAATGGCATCAATTGCGAGCCGGAAGCGGTTAGGGATAACTTTGACTACTTCAACTATGGCGCGAACGATGATGTTTTGAAAAACTTAGAATCTCAATTGTCGAAAGATGAAGTGGTCTTGAGACCTCAAGAGGTCAAAGACTGGCCACAGCAATCTTTGAATGTCGGACAAATAACAGCAGTATCCATTAATTCTTTGGGACAGCCCGTGATATTTCACAGGGCAGATCGAGTATGGGACGAAAATACTTTCAACGAATCCAATGCTTATCAAAACTTCGACAAGGGACCTATAGTTGAAGATACAATTCTTGTTCTTGACCCTGGTAGCGGCTCCGTCCTACATAGCTGGGGAGCTTATATTTTTTATATGCCTCATGGCCTTACCCTCGATCATCATGATAACGTGTGGGTAACTGACGTCGCAAAACATCAAGTATATAAGTATACGCCAAGTAACCACAGATATCCGACCCTGACTATTGGCGAACCCTTCACAGCTGGTCTCCCATTCAGGCATCGTGTTTTATTTTGCATGCCTACATCAGTAGCAATTGCTAGCACCGGAGAAATTTTCGTAGCTGACGGATACTGTAATAATCAAATAGTAAAATTCAATGCCGCTGGGACTCTATTGCTGACGATTCCAGCTTATTCCGACACCTGGTCATTGAATTTGCCTCACAGTGTCACTTTGCTTGAACATTTGGATCTGGTTTGCGTGGCCGATAGAGAAAATATGAGGATCGTGTGCCCGAAGGCTGGATTGAAAAGTTACGCGGATCCGTTAGAGCCACCTACGATAATTGAAGATCCCACTTTGGGCAGGGTGTTTGCAGTGACTTCTCATGGTGATACTATTTACGCTGTGAACGGTCCCACGTCACAGAATATTGCTGTCCGAGGCTTTACTGTTAACGCTGTTTACGGAAACATTTTGGATACTTGGGAACCGACCACTGGATTCACGAACCCACACTCGCTGGCGGTTACAAGGAACGGATCACATCTCTACGTTTCTGAAATTGGACCGAACAAAATCTGGAAGTTTGAATTGACCGACGTATACGATAAGAAATAG
Protein
MLFVLLYAVILNGINCEPEAVRDNFDYFNYGANDDVLKNLESQLSKDEVVLRPQEVKDWPQQSLNVGQITAVSINSLGQPVIFHRADRVWDENTFNESNAYQNFDKGPIVEDTILVLDPGSGSVLHSWGAYIFYMPHGLTLDHHDNVWVTDVAKHQVYKYTPSNHRYPTLTIGEPFTAGLPFRHRVLFCMPTSVAIASTGEIFVADGYCNNQIVKFNAAGTLLLTIPAYSDTWSLNLPHSVTLLEHLDLVCVADRENMRIVCPKAGLKSYADPLEPPTIIEDPTLGRVFAVTSHGDTIYAVNGPTSQNIAVRGFTVNAVYGNILDTWEPTTGFTNPHSLAVTRNGSHLYVSEIGPNKIWKFELTDVYDKK
Summary
Description
Probable lyase that catalyzes an essential reaction in C-terminal alpha-amidation of peptides. Mediates the dismutation of the unstable peptidyl(2-hydroxyglycine) intermediate to glyoxylate and the corresponding desglycine peptide amide. C-terminal amidation of peptides such as neuropeptides is essential for full biological activity.
Catalytic Activity
a [peptide]-C-terminal (2S)-2-hydroxyglycine = a [peptide]-C-terminal amide + glyoxylate
Cofactor
Zn(2+)
Biophysicochemical Properties
70 uM for peptidyl-alpha-hydroxyglycine
Similarity
Belongs to the peptidyl-alpha-hydroxyglycine alpha-amidating lyase family.
Keywords
Complete proteome
Disulfide bond
Glycoprotein
Lyase
Metal-binding
Reference proteome
Repeat
Secreted
Signal
Zinc
Feature
chain Peptidyl-alpha-hydroxyglycine alpha-amidating lyase 2
Uniprot
H9IXC2
A0A194QW59
A0A194QJL1
A0A212F5R2
A0A2H1WAV2
A0A0L7L3L2
+ More
D6WS07 U4UDG4 B4NML5 A0A2J7R7W5 A0A1W4UAC0 A0A182GBJ2 A0A023ERJ6 A0A084WEC6 B3NPQ6 B4P9R3 B4QAX4 B4I8R6 A0A023ESQ7 A0A0B4LG96 Q9W1L5 B4GHU2 A0A182G884 B0WEV2 A0A182NJC7 Q28ZF6 B4KTG3 E0VKF2 A0A1S4G1Z5 A0A023ESU1 B3MFI8 A0A182J0Y5 A0A336N1C7 B4J4G0 A0A336LKU6 A0A034VYR7 A0A3B0J1Z2 A0A2S2QFS7 A0A182MTV0 A0A1Q3FRB9 A0A1Q3FRD4 A0A1Q3FRT6 A0A182FH11 A0A182XMK3 A0A1Q3FRB8 A0A182S982 A0A182TZY5 A0A1Q3FR96 A0A1Q3FRK1 A0A0M4E5T7 Q16GA4 A0A182I2D1 W8AQC3 B4LN29 A0A182WLC9 Q7Q6K9 A0A1B6FIQ8 A0A2M4AY97 A0A182R4J3 A0A182QE76 A0A182PW29 A0A182YEF7 A0A2H8TZW8 A0A1B6I9N2 J9JSH4 A0A0A1WDN3 A0A0A1WE63 W5JAE0 U5ER94 A0A067QWN9 A0A2S2ND71 A0A1B6CDU4 A0A1Y1LW31 A0A1I8MBU3 A0A1B0GBQ4 A0A1I8MBT9 A0A1A9VLB9 A0A1A9YJE4 A0A1B0BZV2 A0A1I8Q792 A0A1A9WQJ0 A0A1A9Z564 A0A0T6BF45 Q171A0 A0A0A9WBR7 N6TA58 A0A026WU18 A0A2P8YNJ4 A0A1W4WYG3 A0A1B6MB47 A0A348G613 A0A1D2N973 A0A226EN09 E2AW67 A0A0P5LX19 A0A0N8AQ75 A0A0P5KUR2 E9HAK8 A0A0P5SHW4 A0A0J7N5A8
D6WS07 U4UDG4 B4NML5 A0A2J7R7W5 A0A1W4UAC0 A0A182GBJ2 A0A023ERJ6 A0A084WEC6 B3NPQ6 B4P9R3 B4QAX4 B4I8R6 A0A023ESQ7 A0A0B4LG96 Q9W1L5 B4GHU2 A0A182G884 B0WEV2 A0A182NJC7 Q28ZF6 B4KTG3 E0VKF2 A0A1S4G1Z5 A0A023ESU1 B3MFI8 A0A182J0Y5 A0A336N1C7 B4J4G0 A0A336LKU6 A0A034VYR7 A0A3B0J1Z2 A0A2S2QFS7 A0A182MTV0 A0A1Q3FRB9 A0A1Q3FRD4 A0A1Q3FRT6 A0A182FH11 A0A182XMK3 A0A1Q3FRB8 A0A182S982 A0A182TZY5 A0A1Q3FR96 A0A1Q3FRK1 A0A0M4E5T7 Q16GA4 A0A182I2D1 W8AQC3 B4LN29 A0A182WLC9 Q7Q6K9 A0A1B6FIQ8 A0A2M4AY97 A0A182R4J3 A0A182QE76 A0A182PW29 A0A182YEF7 A0A2H8TZW8 A0A1B6I9N2 J9JSH4 A0A0A1WDN3 A0A0A1WE63 W5JAE0 U5ER94 A0A067QWN9 A0A2S2ND71 A0A1B6CDU4 A0A1Y1LW31 A0A1I8MBU3 A0A1B0GBQ4 A0A1I8MBT9 A0A1A9VLB9 A0A1A9YJE4 A0A1B0BZV2 A0A1I8Q792 A0A1A9WQJ0 A0A1A9Z564 A0A0T6BF45 Q171A0 A0A0A9WBR7 N6TA58 A0A026WU18 A0A2P8YNJ4 A0A1W4WYG3 A0A1B6MB47 A0A348G613 A0A1D2N973 A0A226EN09 E2AW67 A0A0P5LX19 A0A0N8AQ75 A0A0P5KUR2 E9HAK8 A0A0P5SHW4 A0A0J7N5A8
EC Number
4.3.2.5
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
23537049 17994087 26483478 24945155 24438588 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15198673 15632085 20566863 25348373 17510324 24495485 12364791 14747013 17210077 25244985 25830018 20920257 23761445 24845553 28004739 25315136 25401762 26823975 24508170 30249741 29403074 27289101 20798317 21292972
23537049 17994087 26483478 24945155 24438588 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15198673 15632085 20566863 25348373 17510324 24495485 12364791 14747013 17210077 25244985 25830018 20920257 23761445 24845553 28004739 25315136 25401762 26823975 24508170 30249741 29403074 27289101 20798317 21292972
EMBL
BABH01017977
KQ461073
KPJ09712.1
KQ458575
KPJ05717.1
AGBW02010153
+ More
OWR49064.1 ODYU01007425 SOQ50188.1 JTDY01003185 KOB70005.1 KQ971351 EFA06405.1 KB632085 ERL88641.1 CH964282 EDW85604.1 NEVH01006723 PNF36923.1 JXUM01052604 KQ561742 KXJ77670.1 GAPW01001778 JAC11820.1 ATLV01023201 KE525341 KFB48570.1 CH954179 EDV56847.1 CM000158 EDW92371.1 CM000362 CM002911 EDX08411.1 KMY96086.1 CH480824 EDW56991.1 GAPW01001777 JAC11821.1 AE013599 AHN56571.1 AY007228 AY119165 CH479183 EDW36062.1 JXUM01008139 KQ560254 KXJ83535.1 DS231911 EDS45865.1 CM000071 EAL25657.1 CH933808 EDW08524.1 DS235243 EEB13867.1 GAPW01001240 JAC12358.1 CH902619 EDV36673.1 UFQS01001876 UFQT01001876 SSX12635.1 SSX32078.1 CH916367 EDW01642.1 UFQS01000040 UFQT01000040 SSW98204.1 SSX18590.1 GAKP01010487 GAKP01010486 JAC48466.1 OUUW01000001 SPP72982.1 GGMS01006839 MBY76042.1 AXCM01019187 GFDL01004875 JAV30170.1 GFDL01004865 JAV30180.1 GFDL01004902 JAV30143.1 GFDL01004854 JAV30191.1 GFDL01004874 JAV30171.1 GFDL01004848 JAV30197.1 CP012524 ALC41716.1 CH478302 EAT33269.1 APCN01000146 GAMC01015575 GAMC01015574 GAMC01015573 GAMC01015572 JAB90980.1 CH940648 EDW60033.1 AAAB01008960 EAA11890.4 GECZ01019681 JAS50088.1 GGFK01012237 MBW45558.1 AXCN02000448 GFXV01008038 MBW19843.1 GECU01024061 JAS83645.1 ABLF02036142 ABLF02036149 GBXI01017290 JAC97001.1 GBXI01017095 JAC97196.1 ADMH02002052 ETN59749.1 GANO01002906 JAB56965.1 KK853156 KDR10480.1 GGMR01002448 MBY15067.1 GEDC01025672 JAS11626.1 GEZM01045591 JAV77732.1 CCAG010005864 JXJN01023308 JXJN01023309 LJIG01001067 KRT85873.1 CH477462 EAT40544.1 GBHO01038390 GDHC01012169 JAG05214.1 JAQ06460.1 APGK01045474 APGK01045475 KB741037 ENN74603.1 KK107105 QOIP01000011 EZA59478.1 RLU16802.1 PYGN01000467 PSN45830.1 GEBQ01006822 JAT33155.1 FX985552 BBF97886.1 LJIJ01000138 ODN01798.1 LNIX01000003 OXA58670.1 GL443249 EFN62324.1 GDIQ01188313 JAK63412.1 GDIQ01253659 JAJ98065.1 GDIQ01186760 JAK64965.1 GL732612 EFX71247.1 GDIP01139550 JAL64164.1 LBMM01009775 KMQ87880.1
OWR49064.1 ODYU01007425 SOQ50188.1 JTDY01003185 KOB70005.1 KQ971351 EFA06405.1 KB632085 ERL88641.1 CH964282 EDW85604.1 NEVH01006723 PNF36923.1 JXUM01052604 KQ561742 KXJ77670.1 GAPW01001778 JAC11820.1 ATLV01023201 KE525341 KFB48570.1 CH954179 EDV56847.1 CM000158 EDW92371.1 CM000362 CM002911 EDX08411.1 KMY96086.1 CH480824 EDW56991.1 GAPW01001777 JAC11821.1 AE013599 AHN56571.1 AY007228 AY119165 CH479183 EDW36062.1 JXUM01008139 KQ560254 KXJ83535.1 DS231911 EDS45865.1 CM000071 EAL25657.1 CH933808 EDW08524.1 DS235243 EEB13867.1 GAPW01001240 JAC12358.1 CH902619 EDV36673.1 UFQS01001876 UFQT01001876 SSX12635.1 SSX32078.1 CH916367 EDW01642.1 UFQS01000040 UFQT01000040 SSW98204.1 SSX18590.1 GAKP01010487 GAKP01010486 JAC48466.1 OUUW01000001 SPP72982.1 GGMS01006839 MBY76042.1 AXCM01019187 GFDL01004875 JAV30170.1 GFDL01004865 JAV30180.1 GFDL01004902 JAV30143.1 GFDL01004854 JAV30191.1 GFDL01004874 JAV30171.1 GFDL01004848 JAV30197.1 CP012524 ALC41716.1 CH478302 EAT33269.1 APCN01000146 GAMC01015575 GAMC01015574 GAMC01015573 GAMC01015572 JAB90980.1 CH940648 EDW60033.1 AAAB01008960 EAA11890.4 GECZ01019681 JAS50088.1 GGFK01012237 MBW45558.1 AXCN02000448 GFXV01008038 MBW19843.1 GECU01024061 JAS83645.1 ABLF02036142 ABLF02036149 GBXI01017290 JAC97001.1 GBXI01017095 JAC97196.1 ADMH02002052 ETN59749.1 GANO01002906 JAB56965.1 KK853156 KDR10480.1 GGMR01002448 MBY15067.1 GEDC01025672 JAS11626.1 GEZM01045591 JAV77732.1 CCAG010005864 JXJN01023308 JXJN01023309 LJIG01001067 KRT85873.1 CH477462 EAT40544.1 GBHO01038390 GDHC01012169 JAG05214.1 JAQ06460.1 APGK01045474 APGK01045475 KB741037 ENN74603.1 KK107105 QOIP01000011 EZA59478.1 RLU16802.1 PYGN01000467 PSN45830.1 GEBQ01006822 JAT33155.1 FX985552 BBF97886.1 LJIJ01000138 ODN01798.1 LNIX01000003 OXA58670.1 GL443249 EFN62324.1 GDIQ01188313 JAK63412.1 GDIQ01253659 JAJ98065.1 GDIQ01186760 JAK64965.1 GL732612 EFX71247.1 GDIP01139550 JAL64164.1 LBMM01009775 KMQ87880.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000007266
+ More
UP000030742 UP000007798 UP000235965 UP000192221 UP000069940 UP000249989 UP000030765 UP000008711 UP000002282 UP000000304 UP000001292 UP000000803 UP000008744 UP000002320 UP000075884 UP000001819 UP000009192 UP000009046 UP000007801 UP000075880 UP000001070 UP000268350 UP000075883 UP000069272 UP000076407 UP000075901 UP000075902 UP000092553 UP000008820 UP000075840 UP000008792 UP000075920 UP000007062 UP000075900 UP000075886 UP000075885 UP000076408 UP000007819 UP000000673 UP000027135 UP000095301 UP000092444 UP000078200 UP000092443 UP000092460 UP000095300 UP000091820 UP000092445 UP000019118 UP000053097 UP000279307 UP000245037 UP000192223 UP000094527 UP000198287 UP000000311 UP000000305 UP000036403
UP000030742 UP000007798 UP000235965 UP000192221 UP000069940 UP000249989 UP000030765 UP000008711 UP000002282 UP000000304 UP000001292 UP000000803 UP000008744 UP000002320 UP000075884 UP000001819 UP000009192 UP000009046 UP000007801 UP000075880 UP000001070 UP000268350 UP000075883 UP000069272 UP000076407 UP000075901 UP000075902 UP000092553 UP000008820 UP000075840 UP000008792 UP000075920 UP000007062 UP000075900 UP000075886 UP000075885 UP000076408 UP000007819 UP000000673 UP000027135 UP000095301 UP000092444 UP000078200 UP000092443 UP000092460 UP000095300 UP000091820 UP000092445 UP000019118 UP000053097 UP000279307 UP000245037 UP000192223 UP000094527 UP000198287 UP000000311 UP000000305 UP000036403
Pfam
PF01436 NHL
Interpro
Gene 3D
ProteinModelPortal
H9IXC2
A0A194QW59
A0A194QJL1
A0A212F5R2
A0A2H1WAV2
A0A0L7L3L2
+ More
D6WS07 U4UDG4 B4NML5 A0A2J7R7W5 A0A1W4UAC0 A0A182GBJ2 A0A023ERJ6 A0A084WEC6 B3NPQ6 B4P9R3 B4QAX4 B4I8R6 A0A023ESQ7 A0A0B4LG96 Q9W1L5 B4GHU2 A0A182G884 B0WEV2 A0A182NJC7 Q28ZF6 B4KTG3 E0VKF2 A0A1S4G1Z5 A0A023ESU1 B3MFI8 A0A182J0Y5 A0A336N1C7 B4J4G0 A0A336LKU6 A0A034VYR7 A0A3B0J1Z2 A0A2S2QFS7 A0A182MTV0 A0A1Q3FRB9 A0A1Q3FRD4 A0A1Q3FRT6 A0A182FH11 A0A182XMK3 A0A1Q3FRB8 A0A182S982 A0A182TZY5 A0A1Q3FR96 A0A1Q3FRK1 A0A0M4E5T7 Q16GA4 A0A182I2D1 W8AQC3 B4LN29 A0A182WLC9 Q7Q6K9 A0A1B6FIQ8 A0A2M4AY97 A0A182R4J3 A0A182QE76 A0A182PW29 A0A182YEF7 A0A2H8TZW8 A0A1B6I9N2 J9JSH4 A0A0A1WDN3 A0A0A1WE63 W5JAE0 U5ER94 A0A067QWN9 A0A2S2ND71 A0A1B6CDU4 A0A1Y1LW31 A0A1I8MBU3 A0A1B0GBQ4 A0A1I8MBT9 A0A1A9VLB9 A0A1A9YJE4 A0A1B0BZV2 A0A1I8Q792 A0A1A9WQJ0 A0A1A9Z564 A0A0T6BF45 Q171A0 A0A0A9WBR7 N6TA58 A0A026WU18 A0A2P8YNJ4 A0A1W4WYG3 A0A1B6MB47 A0A348G613 A0A1D2N973 A0A226EN09 E2AW67 A0A0P5LX19 A0A0N8AQ75 A0A0P5KUR2 E9HAK8 A0A0P5SHW4 A0A0J7N5A8
D6WS07 U4UDG4 B4NML5 A0A2J7R7W5 A0A1W4UAC0 A0A182GBJ2 A0A023ERJ6 A0A084WEC6 B3NPQ6 B4P9R3 B4QAX4 B4I8R6 A0A023ESQ7 A0A0B4LG96 Q9W1L5 B4GHU2 A0A182G884 B0WEV2 A0A182NJC7 Q28ZF6 B4KTG3 E0VKF2 A0A1S4G1Z5 A0A023ESU1 B3MFI8 A0A182J0Y5 A0A336N1C7 B4J4G0 A0A336LKU6 A0A034VYR7 A0A3B0J1Z2 A0A2S2QFS7 A0A182MTV0 A0A1Q3FRB9 A0A1Q3FRD4 A0A1Q3FRT6 A0A182FH11 A0A182XMK3 A0A1Q3FRB8 A0A182S982 A0A182TZY5 A0A1Q3FR96 A0A1Q3FRK1 A0A0M4E5T7 Q16GA4 A0A182I2D1 W8AQC3 B4LN29 A0A182WLC9 Q7Q6K9 A0A1B6FIQ8 A0A2M4AY97 A0A182R4J3 A0A182QE76 A0A182PW29 A0A182YEF7 A0A2H8TZW8 A0A1B6I9N2 J9JSH4 A0A0A1WDN3 A0A0A1WE63 W5JAE0 U5ER94 A0A067QWN9 A0A2S2ND71 A0A1B6CDU4 A0A1Y1LW31 A0A1I8MBU3 A0A1B0GBQ4 A0A1I8MBT9 A0A1A9VLB9 A0A1A9YJE4 A0A1B0BZV2 A0A1I8Q792 A0A1A9WQJ0 A0A1A9Z564 A0A0T6BF45 Q171A0 A0A0A9WBR7 N6TA58 A0A026WU18 A0A2P8YNJ4 A0A1W4WYG3 A0A1B6MB47 A0A348G613 A0A1D2N973 A0A226EN09 E2AW67 A0A0P5LX19 A0A0N8AQ75 A0A0P5KUR2 E9HAK8 A0A0P5SHW4 A0A0J7N5A8
PDB
5WM0
E-value=6.27381e-61,
Score=593
Ontologies
GO
Topology
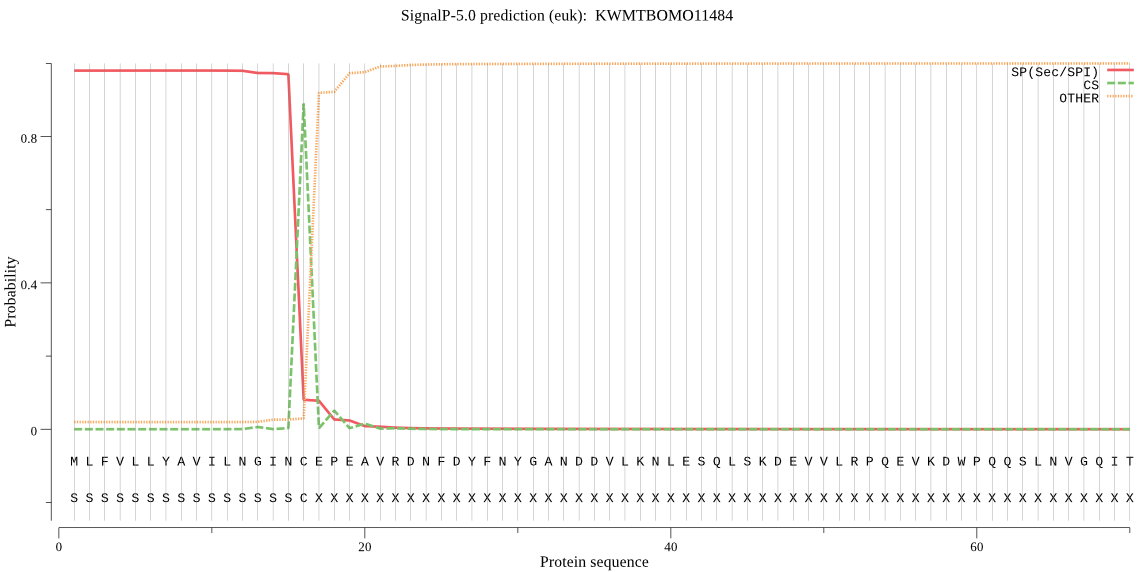
Subcellular location
Secreted
SignalP
Position: 1 - 16,
Likelihood: 0.979757
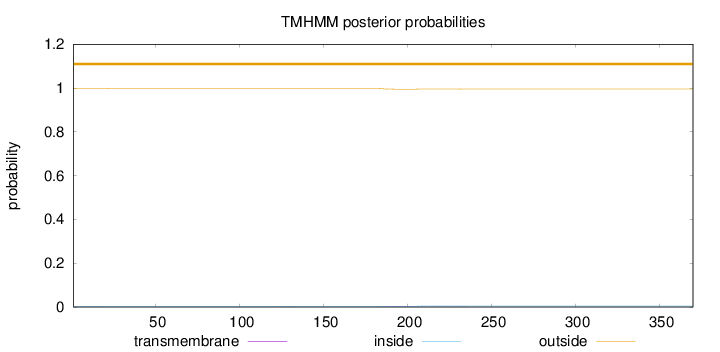
Length:
370
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08981
Exp number, first 60 AAs:
0.01025
Total prob of N-in:
0.00252
outside
1 - 370
Population Genetic Test Statistics
Pi
210.958163
Theta
180.002528
Tajima's D
0.998119
CLR
0.00904
CSRT
0.659567021648918
Interpretation
Uncertain
