Pre Gene Modal
BGIBMGA001918
Annotation
phosphatidylinositol_glycan_precursor_[Bombyx_mori]
Full name
GPI-anchor transamidase
+ More
Putative GPI-anchor transamidase
Putative GPI-anchor transamidase
Alternative Name
Phosphatidylinositol glycan anchor biosynthesis protein class K
Location in the cell
PlasmaMembrane Reliability : 2.45
Sequence
CDS
ATGGCTTTTACAAATCTTATGCTAGTATTCATATTTAATTTATTATATTTATCATTATCATCAGGAATCGAGATACCAGAAGAATTTCAGAAGTCGAATCACACGAATAACTGGGCTGTGCTAGTGGACACTTCACGATTTTGGTTTAACTATCGTCACGTTGCGAATGTTTTATCGATTTATCGGAGTGTGAAACGATTAGGAATACCTGACAGTCAGATTATTCTAATGATTTCTGACGATATGGCATGTAACCCAAGGAATCCCCGACCAGCAACTATTTTTAATAGTGCCCATGAGCAGATCAACGTGTATGGAGATGATGTCGAAGTAGATTATAGAGGTTATGAGGTGTCAGTTGAAAATTTTATCCGTCTGCTAACTGGCAGAGTACCACCTGACACACCTCGCTCCAAGCAATTACTGACTGATGAAGGTAGTAACATACTAATCTACCTAACAGGCCATGGTGGTGACGGCTTCCTCAAGTTTCAGGACTCAGAAGAAGTGACCAGCCAAGAGCTTGCTGATGCTTTGGAACAAATGTGGCAGAAAAAAAGATATAATGAAATTTTTTTCATAATAGACACATGTCAAGCTTCATCAATGTATGAGAAGTTTTATTCACCAAATATATTAGCAACCGCAAGTAGTTTAGTCGGTGAAGATTCACTTTCTCATCATGTTGATTCAGCTATTGGTGTATACATCATTGATAGATACACATACTATGTGCTGGAGTTTTTGGAAAATGTTCACCCTAACAGCAAAAGGACTATGTCGGAATTTTTGGCTGTATGTCCGAAAAGTGCTTGCTTATCGACAGTGGGAGTACGAAAAGATTTATTCAATAGAGATCCTAGTAAAGTGCCTATCACCGACTTCTTTGGTTCAGTTAGACCAGTGATCCTGACAACAGATCCTATAAATATTGTAGAGACACCAAAAAGAAAGAAAAAGACTGAGAATAAAACCAAGATACAAGTGAAACGGGACTATGTGCCACAGTTTCCGATGCATCTAGTCCAAGTTACATAA
Protein
MAFTNLMLVFIFNLLYLSLSSGIEIPEEFQKSNHTNNWAVLVDTSRFWFNYRHVANVLSIYRSVKRLGIPDSQIILMISDDMACNPRNPRPATIFNSAHEQINVYGDDVEVDYRGYEVSVENFIRLLTGRVPPDTPRSKQLLTDEGSNILIYLTGHGGDGFLKFQDSEEVTSQELADALEQMWQKKRYNEIFFIIDTCQASSMYEKFYSPNILATASSLVGEDSLSHHVDSAIGVYIIDRYTYYVLEFLENVHPNSKRTMSEFLAVCPKSACLSTVGVRKDLFNRDPSKVPITDFFGSVRPVILTTDPINIVETPKRKKKTENKTKIQVKRDYVPQFPMHLVQVT
Summary
Description
Mediates GPI anchoring in the endoplasmic reticulum, by replacing a protein's C-terminal GPI attachment signal peptide with a pre-assembled GPI. During this transamidation reaction, the GPI transamidase forms a carbonyl intermediate with the substrate protein.
Mediates GPI anchoring in the endoplasmic reticulum, by replacing a protein's C-terminal GPI attachment signal peptide with a pre-assembled GPI. During this transamidation reaction, the GPI transamidase forms a carbonyl intermediate with the substrate protein (By similarity).
Mediates GPI anchoring in the endoplasmic reticulum, by replacing a protein's C-terminal GPI attachment signal peptide with a pre-assembled GPI. During this transamidation reaction, the GPI transamidase forms a carbonyl intermediate with the substrate protein (By similarity).
Similarity
Belongs to the peptidase C13 family.
Keywords
Complete proteome
GPI-anchor biosynthesis
Hydrolase
Protease
Reference proteome
Signal
Thiol protease
Feature
chain GPI-anchor transamidase
Uniprot
Q1HPH2
A0A2A4JZD3
A0A212FGH5
A0A2H1V6N2
S4PY80
A0A067R5W0
+ More
A0A2P8Y022 A0A088AL45 A0A2J7PSN3 E0VQW5 A0A2A3ERR2 A0A232FIJ4 K7J1G0 A0A0M9A411 A0A1B6MNQ7 H9IXD5 A0A1B6G6C9 A0A0L7QTX1 A0A026VU67 A0A1B6DM73 E2B817 E9J1B2 N6TWV0 A0A0T6ATT9 F4X801 A0A151JAM5 A0A1Y1LJ05 A0A158NVQ9 A0A195FXD0 A0A139WFR6 A0A1Y1LK07 A0A1S3DII2 A0A3Q0JID1 A0A151IDU5 A0A154P611 A0A151XEM8 A0A023F9E8 A0A2R7WAE1 A0A310SKF9 A0A336MQ12 A0A0P4VWX2 R4FLF6 A0A1B0DHL5 A0A0V0GE85 A0A0K8WK85 A0A0A9WQL0 A0A226F1Z0 A0A1J1I510 A0A034WP24 J9JPI1 A0A1W7R826 A0A182G8X8 A0A0L0BVR9 A0A2K8JMF4 A0A1I8N9K8 A0A0P4VZ99 A0A0A1WMN4 A0A1I8NZ40 A0A023EPP1 Q16SV5 A0A0K8TTB9 A0A1L8DU17 A0A1L8DTW9 A0A1W4WJ50 W8C4T3 A0A1L8DU23 A0A182QDG2 A0A182PAI3 A0A1Q3G392 A0A182VY47 A0A1Q3G334 A0A182L6W0 Q7QAC9 A0A182R5M8 A0A151I1X5 A0A182XWR1 E2A5G5 A0A1A9W1Y3 A0A182X8U5 A0A182ID27 A0A1B0BM95 A0A1A9YM54 A0A1A9UGJ1 A0A1B0FAS4 A0A1B0A1V9 A0A2M4DMQ1 A0A2M4ASZ8 B4JK59 B4Q1J6 A0A182F9K1 B3P8U4 A0A2M4ATK0 A0A2M3ZA43 B4I9M5 A0A2M4DMK7 Q8T4E1 A0A2M4BT04 B3MYQ4 B4MES3 A0A3R7SVD3
A0A2P8Y022 A0A088AL45 A0A2J7PSN3 E0VQW5 A0A2A3ERR2 A0A232FIJ4 K7J1G0 A0A0M9A411 A0A1B6MNQ7 H9IXD5 A0A1B6G6C9 A0A0L7QTX1 A0A026VU67 A0A1B6DM73 E2B817 E9J1B2 N6TWV0 A0A0T6ATT9 F4X801 A0A151JAM5 A0A1Y1LJ05 A0A158NVQ9 A0A195FXD0 A0A139WFR6 A0A1Y1LK07 A0A1S3DII2 A0A3Q0JID1 A0A151IDU5 A0A154P611 A0A151XEM8 A0A023F9E8 A0A2R7WAE1 A0A310SKF9 A0A336MQ12 A0A0P4VWX2 R4FLF6 A0A1B0DHL5 A0A0V0GE85 A0A0K8WK85 A0A0A9WQL0 A0A226F1Z0 A0A1J1I510 A0A034WP24 J9JPI1 A0A1W7R826 A0A182G8X8 A0A0L0BVR9 A0A2K8JMF4 A0A1I8N9K8 A0A0P4VZ99 A0A0A1WMN4 A0A1I8NZ40 A0A023EPP1 Q16SV5 A0A0K8TTB9 A0A1L8DU17 A0A1L8DTW9 A0A1W4WJ50 W8C4T3 A0A1L8DU23 A0A182QDG2 A0A182PAI3 A0A1Q3G392 A0A182VY47 A0A1Q3G334 A0A182L6W0 Q7QAC9 A0A182R5M8 A0A151I1X5 A0A182XWR1 E2A5G5 A0A1A9W1Y3 A0A182X8U5 A0A182ID27 A0A1B0BM95 A0A1A9YM54 A0A1A9UGJ1 A0A1B0FAS4 A0A1B0A1V9 A0A2M4DMQ1 A0A2M4ASZ8 B4JK59 B4Q1J6 A0A182F9K1 B3P8U4 A0A2M4ATK0 A0A2M3ZA43 B4I9M5 A0A2M4DMK7 Q8T4E1 A0A2M4BT04 B3MYQ4 B4MES3 A0A3R7SVD3
EC Number
3.-.-.-
Pubmed
22118469
23622113
24845553
29403074
20566863
28648823
+ More
20075255 19121390 24508170 30249741 20798317 21282665 23537049 21719571 28004739 21347285 18362917 19820115 25474469 27129103 25401762 26823975 25348373 26483478 26108605 25315136 25830018 24945155 17510324 26369729 24495485 20966253 12364791 14747013 17210077 25244985 17994087 17550304 10731132 12537572 10731137 12537569 18057021
20075255 19121390 24508170 30249741 20798317 21282665 23537049 21719571 28004739 21347285 18362917 19820115 25474469 27129103 25401762 26823975 25348373 26483478 26108605 25315136 25830018 24945155 17510324 26369729 24495485 20966253 12364791 14747013 17210077 25244985 17994087 17550304 10731132 12537572 10731137 12537569 18057021
EMBL
DQ443430
ABF51519.1
NWSH01000386
PCG76770.1
AGBW02008683
OWR52803.1
+ More
ODYU01000957 SOQ36498.1 GAIX01003753 JAA88807.1 KK852722 KDR17719.1 PYGN01001100 PSN37596.1 NEVH01021925 PNF19345.1 DS235442 EEB15771.1 KZ288191 PBC34455.1 NNAY01000193 OXU30117.1 KQ435741 KOX76767.1 GEBQ01002434 JAT37543.1 BABH01017977 GECZ01011773 JAS57996.1 KQ414740 KOC62070.1 KK107899 QOIP01000006 EZA47277.1 RLU21554.1 GEDC01010550 JAS26748.1 GL446273 EFN88159.1 GL767601 EFZ13391.1 APGK01057802 KB741282 KB631802 ENN70787.1 ERL86239.1 LJIG01022832 KRT78488.1 GL888900 EGI57418.1 KQ979294 KYN22003.1 GEZM01058070 JAV71925.1 ADTU01027401 KQ981193 KYN45093.1 KQ971352 KYB26647.1 GEZM01058069 JAV71926.1 KQ977907 KYM98840.1 KQ434809 KZC06638.1 KQ982249 KYQ58829.1 GBBI01001088 JAC17624.1 KK854536 PTY16654.1 KQ761917 OAD56436.1 UFQS01001999 UFQT01001999 SSX12914.1 SSX32356.1 GDKW01002511 JAI54084.1 ACPB03008305 GAHY01002129 JAA75381.1 AJVK01061283 GECL01000514 JAP05610.1 GDHF01032811 GDHF01002800 GDHF01000812 JAI19503.1 JAI49514.1 JAI51502.1 GBHO01036513 GBHO01036509 GBHO01036508 GBRD01014348 GBRD01004175 GDHC01020969 GDHC01013854 JAG07091.1 JAG07095.1 JAG07096.1 JAG51478.1 JAP97659.1 JAQ04775.1 LNIX01000001 OXA63799.1 CVRI01000037 CRK93465.1 GAKP01002557 GAKP01002555 GAKP01002553 GAKP01002551 JAC56399.1 ABLF02037822 GEHC01000382 JAV47263.1 JXUM01048525 JXUM01048526 JXUM01048527 KQ561566 KXJ78150.1 JRES01001264 KNC24093.1 KY031103 ATU82854.1 GDRN01101436 JAI58377.1 GBXI01014371 JAC99920.1 GAPW01002116 JAC11482.1 CH477664 EAT37564.1 GDAI01000428 JAI17175.1 GFDF01004194 JAV09890.1 GFDF01004193 JAV09891.1 GAMC01005294 JAC01262.1 GFDF01004180 JAV09904.1 AXCN02001189 GFDL01000836 JAV34209.1 GFDL01000835 JAV34210.1 AAAB01008898 EAA09153.5 KQ976554 KYM80804.1 GL436931 EFN71330.1 APCN01004978 JXJN01016746 CCAG010005731 GGFL01014627 MBW78805.1 GGFK01010588 MBW43909.1 CH916370 EDV99961.1 CM000162 EDX01437.1 CH954183 EDV45549.1 GGFK01010587 MBW43908.1 GGFM01004621 MBW25372.1 CH480825 EDW43906.1 GGFL01014628 MBW78806.1 AE014298 AL009192 AY089234 AAF45703.2 AAL89972.1 CAA15687.1 GGFJ01007001 MBW56142.1 CH902632 EDV32748.1 CH940664 EDW63048.1 KRF80881.1 QCYY01001598 ROT76880.1
ODYU01000957 SOQ36498.1 GAIX01003753 JAA88807.1 KK852722 KDR17719.1 PYGN01001100 PSN37596.1 NEVH01021925 PNF19345.1 DS235442 EEB15771.1 KZ288191 PBC34455.1 NNAY01000193 OXU30117.1 KQ435741 KOX76767.1 GEBQ01002434 JAT37543.1 BABH01017977 GECZ01011773 JAS57996.1 KQ414740 KOC62070.1 KK107899 QOIP01000006 EZA47277.1 RLU21554.1 GEDC01010550 JAS26748.1 GL446273 EFN88159.1 GL767601 EFZ13391.1 APGK01057802 KB741282 KB631802 ENN70787.1 ERL86239.1 LJIG01022832 KRT78488.1 GL888900 EGI57418.1 KQ979294 KYN22003.1 GEZM01058070 JAV71925.1 ADTU01027401 KQ981193 KYN45093.1 KQ971352 KYB26647.1 GEZM01058069 JAV71926.1 KQ977907 KYM98840.1 KQ434809 KZC06638.1 KQ982249 KYQ58829.1 GBBI01001088 JAC17624.1 KK854536 PTY16654.1 KQ761917 OAD56436.1 UFQS01001999 UFQT01001999 SSX12914.1 SSX32356.1 GDKW01002511 JAI54084.1 ACPB03008305 GAHY01002129 JAA75381.1 AJVK01061283 GECL01000514 JAP05610.1 GDHF01032811 GDHF01002800 GDHF01000812 JAI19503.1 JAI49514.1 JAI51502.1 GBHO01036513 GBHO01036509 GBHO01036508 GBRD01014348 GBRD01004175 GDHC01020969 GDHC01013854 JAG07091.1 JAG07095.1 JAG07096.1 JAG51478.1 JAP97659.1 JAQ04775.1 LNIX01000001 OXA63799.1 CVRI01000037 CRK93465.1 GAKP01002557 GAKP01002555 GAKP01002553 GAKP01002551 JAC56399.1 ABLF02037822 GEHC01000382 JAV47263.1 JXUM01048525 JXUM01048526 JXUM01048527 KQ561566 KXJ78150.1 JRES01001264 KNC24093.1 KY031103 ATU82854.1 GDRN01101436 JAI58377.1 GBXI01014371 JAC99920.1 GAPW01002116 JAC11482.1 CH477664 EAT37564.1 GDAI01000428 JAI17175.1 GFDF01004194 JAV09890.1 GFDF01004193 JAV09891.1 GAMC01005294 JAC01262.1 GFDF01004180 JAV09904.1 AXCN02001189 GFDL01000836 JAV34209.1 GFDL01000835 JAV34210.1 AAAB01008898 EAA09153.5 KQ976554 KYM80804.1 GL436931 EFN71330.1 APCN01004978 JXJN01016746 CCAG010005731 GGFL01014627 MBW78805.1 GGFK01010588 MBW43909.1 CH916370 EDV99961.1 CM000162 EDX01437.1 CH954183 EDV45549.1 GGFK01010587 MBW43908.1 GGFM01004621 MBW25372.1 CH480825 EDW43906.1 GGFL01014628 MBW78806.1 AE014298 AL009192 AY089234 AAF45703.2 AAL89972.1 CAA15687.1 GGFJ01007001 MBW56142.1 CH902632 EDV32748.1 CH940664 EDW63048.1 KRF80881.1 QCYY01001598 ROT76880.1
Proteomes
UP000218220
UP000007151
UP000027135
UP000245037
UP000005203
UP000235965
+ More
UP000009046 UP000242457 UP000215335 UP000002358 UP000053105 UP000005204 UP000053825 UP000053097 UP000279307 UP000008237 UP000019118 UP000030742 UP000007755 UP000078492 UP000005205 UP000078541 UP000007266 UP000079169 UP000078542 UP000076502 UP000075809 UP000015103 UP000092462 UP000198287 UP000183832 UP000007819 UP000069940 UP000249989 UP000037069 UP000095301 UP000095300 UP000008820 UP000192223 UP000075886 UP000075885 UP000075920 UP000075882 UP000007062 UP000075900 UP000078540 UP000076408 UP000000311 UP000091820 UP000076407 UP000075840 UP000092460 UP000092443 UP000078200 UP000092444 UP000092445 UP000001070 UP000002282 UP000069272 UP000008711 UP000001292 UP000000803 UP000007801 UP000008792 UP000283509
UP000009046 UP000242457 UP000215335 UP000002358 UP000053105 UP000005204 UP000053825 UP000053097 UP000279307 UP000008237 UP000019118 UP000030742 UP000007755 UP000078492 UP000005205 UP000078541 UP000007266 UP000079169 UP000078542 UP000076502 UP000075809 UP000015103 UP000092462 UP000198287 UP000183832 UP000007819 UP000069940 UP000249989 UP000037069 UP000095301 UP000095300 UP000008820 UP000192223 UP000075886 UP000075885 UP000075920 UP000075882 UP000007062 UP000075900 UP000078540 UP000076408 UP000000311 UP000091820 UP000076407 UP000075840 UP000092460 UP000092443 UP000078200 UP000092444 UP000092445 UP000001070 UP000002282 UP000069272 UP000008711 UP000001292 UP000000803 UP000007801 UP000008792 UP000283509
Interpro
Gene 3D
ProteinModelPortal
Q1HPH2
A0A2A4JZD3
A0A212FGH5
A0A2H1V6N2
S4PY80
A0A067R5W0
+ More
A0A2P8Y022 A0A088AL45 A0A2J7PSN3 E0VQW5 A0A2A3ERR2 A0A232FIJ4 K7J1G0 A0A0M9A411 A0A1B6MNQ7 H9IXD5 A0A1B6G6C9 A0A0L7QTX1 A0A026VU67 A0A1B6DM73 E2B817 E9J1B2 N6TWV0 A0A0T6ATT9 F4X801 A0A151JAM5 A0A1Y1LJ05 A0A158NVQ9 A0A195FXD0 A0A139WFR6 A0A1Y1LK07 A0A1S3DII2 A0A3Q0JID1 A0A151IDU5 A0A154P611 A0A151XEM8 A0A023F9E8 A0A2R7WAE1 A0A310SKF9 A0A336MQ12 A0A0P4VWX2 R4FLF6 A0A1B0DHL5 A0A0V0GE85 A0A0K8WK85 A0A0A9WQL0 A0A226F1Z0 A0A1J1I510 A0A034WP24 J9JPI1 A0A1W7R826 A0A182G8X8 A0A0L0BVR9 A0A2K8JMF4 A0A1I8N9K8 A0A0P4VZ99 A0A0A1WMN4 A0A1I8NZ40 A0A023EPP1 Q16SV5 A0A0K8TTB9 A0A1L8DU17 A0A1L8DTW9 A0A1W4WJ50 W8C4T3 A0A1L8DU23 A0A182QDG2 A0A182PAI3 A0A1Q3G392 A0A182VY47 A0A1Q3G334 A0A182L6W0 Q7QAC9 A0A182R5M8 A0A151I1X5 A0A182XWR1 E2A5G5 A0A1A9W1Y3 A0A182X8U5 A0A182ID27 A0A1B0BM95 A0A1A9YM54 A0A1A9UGJ1 A0A1B0FAS4 A0A1B0A1V9 A0A2M4DMQ1 A0A2M4ASZ8 B4JK59 B4Q1J6 A0A182F9K1 B3P8U4 A0A2M4ATK0 A0A2M3ZA43 B4I9M5 A0A2M4DMK7 Q8T4E1 A0A2M4BT04 B3MYQ4 B4MES3 A0A3R7SVD3
A0A2P8Y022 A0A088AL45 A0A2J7PSN3 E0VQW5 A0A2A3ERR2 A0A232FIJ4 K7J1G0 A0A0M9A411 A0A1B6MNQ7 H9IXD5 A0A1B6G6C9 A0A0L7QTX1 A0A026VU67 A0A1B6DM73 E2B817 E9J1B2 N6TWV0 A0A0T6ATT9 F4X801 A0A151JAM5 A0A1Y1LJ05 A0A158NVQ9 A0A195FXD0 A0A139WFR6 A0A1Y1LK07 A0A1S3DII2 A0A3Q0JID1 A0A151IDU5 A0A154P611 A0A151XEM8 A0A023F9E8 A0A2R7WAE1 A0A310SKF9 A0A336MQ12 A0A0P4VWX2 R4FLF6 A0A1B0DHL5 A0A0V0GE85 A0A0K8WK85 A0A0A9WQL0 A0A226F1Z0 A0A1J1I510 A0A034WP24 J9JPI1 A0A1W7R826 A0A182G8X8 A0A0L0BVR9 A0A2K8JMF4 A0A1I8N9K8 A0A0P4VZ99 A0A0A1WMN4 A0A1I8NZ40 A0A023EPP1 Q16SV5 A0A0K8TTB9 A0A1L8DU17 A0A1L8DTW9 A0A1W4WJ50 W8C4T3 A0A1L8DU23 A0A182QDG2 A0A182PAI3 A0A1Q3G392 A0A182VY47 A0A1Q3G334 A0A182L6W0 Q7QAC9 A0A182R5M8 A0A151I1X5 A0A182XWR1 E2A5G5 A0A1A9W1Y3 A0A182X8U5 A0A182ID27 A0A1B0BM95 A0A1A9YM54 A0A1A9UGJ1 A0A1B0FAS4 A0A1B0A1V9 A0A2M4DMQ1 A0A2M4ASZ8 B4JK59 B4Q1J6 A0A182F9K1 B3P8U4 A0A2M4ATK0 A0A2M3ZA43 B4I9M5 A0A2M4DMK7 Q8T4E1 A0A2M4BT04 B3MYQ4 B4MES3 A0A3R7SVD3
PDB
4NOM
E-value=2.05938e-25,
Score=286
Ontologies
PATHWAY
GO
PANTHER
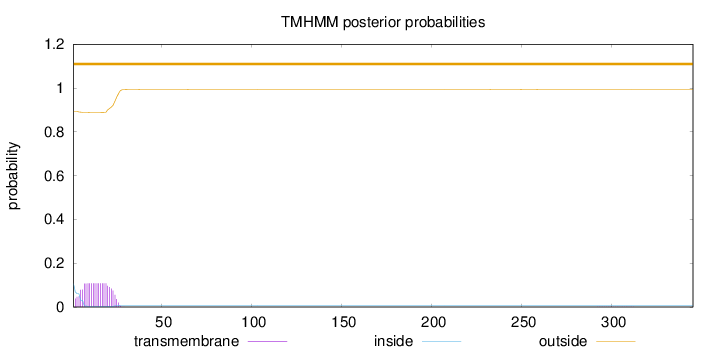
Topology
SignalP
Position: 1 - 22,
Likelihood: 0.978711
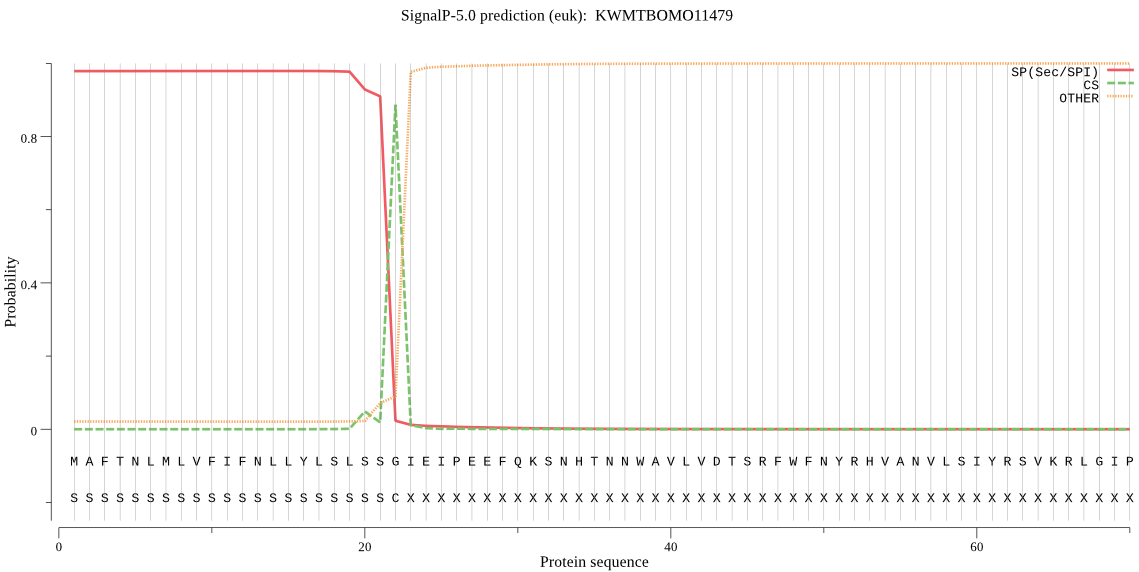
Length:
345
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.16720000000001
Exp number, first 60 AAs:
2.16273
Total prob of N-in:
0.10697
outside
1 - 345
Population Genetic Test Statistics
Pi
152.577554
Theta
162.037086
Tajima's D
-0.470052
CLR
0.598643
CSRT
0.248037598120094
Interpretation
Uncertain
