Gene
KWMTBOMO11468
Pre Gene Modal
BGIBMGA001920
Annotation
PREDICTED:_leptin_receptor_gene-related_protein_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.943
Sequence
CDS
ATGGACACAATAGGTTTGGTGTCGTTGGCATTCGCGGGTTCAATTGGAATGACATTTGTCATACTAGCTTGTGCCTTACCACAATACAAACTATGGTGGCCATTCTTCGTTGTGCTATTCTACATTCTGTGCCCAATACCTACGATGATCGCTAGGAGGCATACTGATGGTGCTGGAGGTTCAAATTCCGCTTGTATGGAAACTGCAGTATTTATCACAATGGGATTTTTAGTCAGCTCTTTTGCGCTACCGATAGTTCTCGCCAGGGCTGGTGTGATTTTTTGGGGTGCATGTTACCTTACACTGGCCGGTAACGTGATCGTATATCTGACGATCTTGGGTTTCTTCACCATCTTTGACATGGACGACTCCGATTATGCGATGTGGTGA
Protein
MDTIGLVSLAFAGSIGMTFVILACALPQYKLWWPFFVVLFYILCPIPTMIARRHTDGAGGSNSACMETAVFITMGFLVSSFALPIVLARAGVIFWGACYLTLAGNVIVYLTILGFFTIFDMDDSDYAMW
Summary
Uniprot
S4NWK7
A0A2H1X1P6
I4DJ64
A0A142LUZ6
A0A194QXN9
A0A1E1WI25
+ More
A0A212EPV2 A0A3S2NTP2 A0A194QL92 A0A2A4JLB4 A0A1W4WK96 E2AP43 F4W4F5 A0A195EZ52 A0A158P374 J3JXR5 E2BAB0 R4V4M2 A0A0C9R707 A0A195DMX4 A0A151WYZ2 A0A195BIR9 A0A139WFE7 K7IY04 A0A232F276 A0A1L8DK75 A0A026W241 A0A1B6I8Q9 A0A195CF90 R4WDU1 R4G3W8 A0A224Y169 A0A1B6MAZ1 A0A1B0GGT7 A0A0P4VNL2 Q7YSY6 A0A023FAG5 A0A1B6LAF9 A0A0V0G572 A0A1B6CTT9 N6U7F3 A0A1B6DPA8 A0A2P8YSZ3 A0A0C9RA89 A0A0M9A0G5 A0A0A9XZF6 A0A1B6FRV0 A0A1B6CXD3 C4WSX4 A0A0P4VZJ7 A0A210PNU1 E9K6D5 T1HVL4 A0A0P6HW74 J9K6V6 E9H1B9 A0A0N8D1I6 A0A2R5LFE8 A0A2H8TJH4 A0A2T7NNE3 A0A0P6ASI4 A0A2S2R7S4 A0A1Q3FJB3 A0A131ZBC3 A0A224YL29 B0W1T5 G3MKX9 A0A023FLN2 A0A023FTP3 Q16G90 A0A2J7R5F5 C3YTG4 A0A023EF32 A0A3S3SMC1 U5EWY2 A0A2L2YA92 A0A3Q3KTS6 A0A3Q3KH25 L7LUW6 A0A087TSL1 A0A2L2YA94 A0A0L8G645 V5IIH3 T1JGY5 A0A3Q3KJ66 A0A131Y7F2 V5HVW0 A0A2A3EAY5 A0A3B4YL21 A0A3B4TWL8 A0A2U9BK68 W5J9F6 Q7SZ72 A0A182R7Y6 A0A182F3A2 A0A091NKG5 A0A131YBU8 A0A093I3Y3
A0A212EPV2 A0A3S2NTP2 A0A194QL92 A0A2A4JLB4 A0A1W4WK96 E2AP43 F4W4F5 A0A195EZ52 A0A158P374 J3JXR5 E2BAB0 R4V4M2 A0A0C9R707 A0A195DMX4 A0A151WYZ2 A0A195BIR9 A0A139WFE7 K7IY04 A0A232F276 A0A1L8DK75 A0A026W241 A0A1B6I8Q9 A0A195CF90 R4WDU1 R4G3W8 A0A224Y169 A0A1B6MAZ1 A0A1B0GGT7 A0A0P4VNL2 Q7YSY6 A0A023FAG5 A0A1B6LAF9 A0A0V0G572 A0A1B6CTT9 N6U7F3 A0A1B6DPA8 A0A2P8YSZ3 A0A0C9RA89 A0A0M9A0G5 A0A0A9XZF6 A0A1B6FRV0 A0A1B6CXD3 C4WSX4 A0A0P4VZJ7 A0A210PNU1 E9K6D5 T1HVL4 A0A0P6HW74 J9K6V6 E9H1B9 A0A0N8D1I6 A0A2R5LFE8 A0A2H8TJH4 A0A2T7NNE3 A0A0P6ASI4 A0A2S2R7S4 A0A1Q3FJB3 A0A131ZBC3 A0A224YL29 B0W1T5 G3MKX9 A0A023FLN2 A0A023FTP3 Q16G90 A0A2J7R5F5 C3YTG4 A0A023EF32 A0A3S3SMC1 U5EWY2 A0A2L2YA92 A0A3Q3KTS6 A0A3Q3KH25 L7LUW6 A0A087TSL1 A0A2L2YA94 A0A0L8G645 V5IIH3 T1JGY5 A0A3Q3KJ66 A0A131Y7F2 V5HVW0 A0A2A3EAY5 A0A3B4YL21 A0A3B4TWL8 A0A2U9BK68 W5J9F6 Q7SZ72 A0A182R7Y6 A0A182F3A2 A0A091NKG5 A0A131YBU8 A0A093I3Y3
Pubmed
23622113
22651552
26354079
22118469
20798317
21719571
+ More
21347285 22516182 18362917 19820115 20075255 28648823 24508170 23691247 27129103 14976983 25474469 23537049 29403074 25401762 26823975 28812685 20567508 21292972 26830274 28797301 22216098 17510324 18563158 24945155 26483478 26561354 25576852 25765539 20920257 23761445
21347285 22516182 18362917 19820115 20075255 28648823 24508170 23691247 27129103 14976983 25474469 23537049 29403074 25401762 26823975 28812685 20567508 21292972 26830274 28797301 22216098 17510324 18563158 24945155 26483478 26561354 25576852 25765539 20920257 23761445
EMBL
GAIX01009419
JAA83141.1
ODYU01012730
SOQ59182.1
AK401332
BAM17954.1
+ More
KT963072 AMS37611.1 KQ461073 KPJ09725.1 GDQN01004422 GDQN01003469 JAT86632.1 JAT87585.1 AGBW02013366 OWR43518.1 RSAL01000086 RVE48302.1 KQ458575 KPJ05705.1 NWSH01001178 PCG72233.1 GL441452 EFN64779.1 GL887532 EGI70850.1 KQ981905 KYN33585.1 ADTU01007770 BT128036 AEE62997.1 GL446696 EFN87365.1 KC740989 AGM32813.1 GBYB01003725 JAG73492.1 KQ980713 KYN14255.1 KQ982649 KYQ53103.1 KQ976467 KYM84185.1 KQ971352 KYB26652.1 NNAY01001254 OXU24603.1 GFDF01007334 JAV06750.1 KK107483 EZA50137.1 GECU01024417 JAS83289.1 KQ977935 KYM98708.1 AK417968 BAN21183.1 GAHY01001855 JAA75655.1 GFTR01002247 JAW14179.1 GEBQ01006884 JAT33093.1 AJWK01000737 GDKW01002066 JAI54529.1 AY340276 AAQ20841.1 GBBI01000713 JAC17999.1 GEBQ01019483 JAT20494.1 GECL01002929 JAP03195.1 GEDC01020481 JAS16817.1 APGK01045788 KB741039 KB632367 ENN74522.1 ERL93661.1 GEDC01009797 JAS27501.1 PYGN01000378 PSN47355.1 GBYB01003726 JAG73493.1 KQ435800 KOX73299.1 GBHO01018828 GDHC01009170 JAG24776.1 JAQ09459.1 GECZ01016875 JAS52894.1 GEDC01019181 JAS18117.1 AK340371 BAH70994.1 GDRN01095874 JAI59447.1 NEDP02005572 OWF38175.1 GU443952 GU443953 ADV57398.1 ACPB03010726 ACPB03010727 ACPB03010728 ACPB03010729 ACPB03010730 ACPB03010731 ACPB03010732 ACPB03010733 GDIQ01013374 JAN81363.1 ABLF02038030 GL732583 EFX74446.1 GDIQ01134706 GDIP01077959 JAL17020.1 JAM25756.1 GGLE01004052 MBY08178.1 GFXV01001633 MBW13438.1 PZQS01000010 PVD22691.1 GDIP01025025 JAM78690.1 GGMS01016868 MBY86071.1 GFDL01007430 JAV27615.1 GEDV01000771 JAP87786.1 GFPF01003787 MAA14933.1 DS231823 EDS26636.1 JO842530 AEO34147.1 GBBK01002005 JAC22477.1 GBBL01002273 JAC25047.1 CH478310 EAT33258.1 NEVH01006994 PNF36063.1 GG666551 EEN56531.1 JXUM01060121 GAPW01006200 GAPW01006199 GAPW01006198 KQ562088 JAC07400.1 KXJ76721.1 NCKU01000114 RWS17152.1 GANO01002766 JAB57105.1 IAAA01010099 LAA05064.1 GACK01010351 JAA54683.1 KK116552 KFM68100.1 IAAA01010100 LAA05068.1 KQ423631 KOF72511.1 GANP01003187 JAB81281.1 JH432213 GEFM01000637 JAP75159.1 GANP01001724 JAB82744.1 KZ288301 PBC28850.1 CP026249 AWP04454.1 ADMH02001784 ETN61092.1 BC053822 AAH53822.1 KL384943 KFP89562.1 GEFM01000375 JAP75421.1 KL214744 KFV61481.1
KT963072 AMS37611.1 KQ461073 KPJ09725.1 GDQN01004422 GDQN01003469 JAT86632.1 JAT87585.1 AGBW02013366 OWR43518.1 RSAL01000086 RVE48302.1 KQ458575 KPJ05705.1 NWSH01001178 PCG72233.1 GL441452 EFN64779.1 GL887532 EGI70850.1 KQ981905 KYN33585.1 ADTU01007770 BT128036 AEE62997.1 GL446696 EFN87365.1 KC740989 AGM32813.1 GBYB01003725 JAG73492.1 KQ980713 KYN14255.1 KQ982649 KYQ53103.1 KQ976467 KYM84185.1 KQ971352 KYB26652.1 NNAY01001254 OXU24603.1 GFDF01007334 JAV06750.1 KK107483 EZA50137.1 GECU01024417 JAS83289.1 KQ977935 KYM98708.1 AK417968 BAN21183.1 GAHY01001855 JAA75655.1 GFTR01002247 JAW14179.1 GEBQ01006884 JAT33093.1 AJWK01000737 GDKW01002066 JAI54529.1 AY340276 AAQ20841.1 GBBI01000713 JAC17999.1 GEBQ01019483 JAT20494.1 GECL01002929 JAP03195.1 GEDC01020481 JAS16817.1 APGK01045788 KB741039 KB632367 ENN74522.1 ERL93661.1 GEDC01009797 JAS27501.1 PYGN01000378 PSN47355.1 GBYB01003726 JAG73493.1 KQ435800 KOX73299.1 GBHO01018828 GDHC01009170 JAG24776.1 JAQ09459.1 GECZ01016875 JAS52894.1 GEDC01019181 JAS18117.1 AK340371 BAH70994.1 GDRN01095874 JAI59447.1 NEDP02005572 OWF38175.1 GU443952 GU443953 ADV57398.1 ACPB03010726 ACPB03010727 ACPB03010728 ACPB03010729 ACPB03010730 ACPB03010731 ACPB03010732 ACPB03010733 GDIQ01013374 JAN81363.1 ABLF02038030 GL732583 EFX74446.1 GDIQ01134706 GDIP01077959 JAL17020.1 JAM25756.1 GGLE01004052 MBY08178.1 GFXV01001633 MBW13438.1 PZQS01000010 PVD22691.1 GDIP01025025 JAM78690.1 GGMS01016868 MBY86071.1 GFDL01007430 JAV27615.1 GEDV01000771 JAP87786.1 GFPF01003787 MAA14933.1 DS231823 EDS26636.1 JO842530 AEO34147.1 GBBK01002005 JAC22477.1 GBBL01002273 JAC25047.1 CH478310 EAT33258.1 NEVH01006994 PNF36063.1 GG666551 EEN56531.1 JXUM01060121 GAPW01006200 GAPW01006199 GAPW01006198 KQ562088 JAC07400.1 KXJ76721.1 NCKU01000114 RWS17152.1 GANO01002766 JAB57105.1 IAAA01010099 LAA05064.1 GACK01010351 JAA54683.1 KK116552 KFM68100.1 IAAA01010100 LAA05068.1 KQ423631 KOF72511.1 GANP01003187 JAB81281.1 JH432213 GEFM01000637 JAP75159.1 GANP01001724 JAB82744.1 KZ288301 PBC28850.1 CP026249 AWP04454.1 ADMH02001784 ETN61092.1 BC053822 AAH53822.1 KL384943 KFP89562.1 GEFM01000375 JAP75421.1 KL214744 KFV61481.1
Proteomes
UP000053240
UP000007151
UP000283053
UP000053268
UP000218220
UP000192223
+ More
UP000000311 UP000007755 UP000078541 UP000005205 UP000008237 UP000078492 UP000075809 UP000078540 UP000007266 UP000002358 UP000215335 UP000053097 UP000078542 UP000092461 UP000019118 UP000030742 UP000245037 UP000053105 UP000242188 UP000015103 UP000007819 UP000000305 UP000245119 UP000002320 UP000008820 UP000235965 UP000001554 UP000069940 UP000249989 UP000285301 UP000261640 UP000054359 UP000053454 UP000242457 UP000261360 UP000261420 UP000246464 UP000000673 UP000075900 UP000069272 UP000053875
UP000000311 UP000007755 UP000078541 UP000005205 UP000008237 UP000078492 UP000075809 UP000078540 UP000007266 UP000002358 UP000215335 UP000053097 UP000078542 UP000092461 UP000019118 UP000030742 UP000245037 UP000053105 UP000242188 UP000015103 UP000007819 UP000000305 UP000245119 UP000002320 UP000008820 UP000235965 UP000001554 UP000069940 UP000249989 UP000285301 UP000261640 UP000054359 UP000053454 UP000242457 UP000261360 UP000261420 UP000246464 UP000000673 UP000075900 UP000069272 UP000053875
Pfam
PF04133 Vps55
Interpro
IPR007262
Vps55/LEPROT
ProteinModelPortal
S4NWK7
A0A2H1X1P6
I4DJ64
A0A142LUZ6
A0A194QXN9
A0A1E1WI25
+ More
A0A212EPV2 A0A3S2NTP2 A0A194QL92 A0A2A4JLB4 A0A1W4WK96 E2AP43 F4W4F5 A0A195EZ52 A0A158P374 J3JXR5 E2BAB0 R4V4M2 A0A0C9R707 A0A195DMX4 A0A151WYZ2 A0A195BIR9 A0A139WFE7 K7IY04 A0A232F276 A0A1L8DK75 A0A026W241 A0A1B6I8Q9 A0A195CF90 R4WDU1 R4G3W8 A0A224Y169 A0A1B6MAZ1 A0A1B0GGT7 A0A0P4VNL2 Q7YSY6 A0A023FAG5 A0A1B6LAF9 A0A0V0G572 A0A1B6CTT9 N6U7F3 A0A1B6DPA8 A0A2P8YSZ3 A0A0C9RA89 A0A0M9A0G5 A0A0A9XZF6 A0A1B6FRV0 A0A1B6CXD3 C4WSX4 A0A0P4VZJ7 A0A210PNU1 E9K6D5 T1HVL4 A0A0P6HW74 J9K6V6 E9H1B9 A0A0N8D1I6 A0A2R5LFE8 A0A2H8TJH4 A0A2T7NNE3 A0A0P6ASI4 A0A2S2R7S4 A0A1Q3FJB3 A0A131ZBC3 A0A224YL29 B0W1T5 G3MKX9 A0A023FLN2 A0A023FTP3 Q16G90 A0A2J7R5F5 C3YTG4 A0A023EF32 A0A3S3SMC1 U5EWY2 A0A2L2YA92 A0A3Q3KTS6 A0A3Q3KH25 L7LUW6 A0A087TSL1 A0A2L2YA94 A0A0L8G645 V5IIH3 T1JGY5 A0A3Q3KJ66 A0A131Y7F2 V5HVW0 A0A2A3EAY5 A0A3B4YL21 A0A3B4TWL8 A0A2U9BK68 W5J9F6 Q7SZ72 A0A182R7Y6 A0A182F3A2 A0A091NKG5 A0A131YBU8 A0A093I3Y3
A0A212EPV2 A0A3S2NTP2 A0A194QL92 A0A2A4JLB4 A0A1W4WK96 E2AP43 F4W4F5 A0A195EZ52 A0A158P374 J3JXR5 E2BAB0 R4V4M2 A0A0C9R707 A0A195DMX4 A0A151WYZ2 A0A195BIR9 A0A139WFE7 K7IY04 A0A232F276 A0A1L8DK75 A0A026W241 A0A1B6I8Q9 A0A195CF90 R4WDU1 R4G3W8 A0A224Y169 A0A1B6MAZ1 A0A1B0GGT7 A0A0P4VNL2 Q7YSY6 A0A023FAG5 A0A1B6LAF9 A0A0V0G572 A0A1B6CTT9 N6U7F3 A0A1B6DPA8 A0A2P8YSZ3 A0A0C9RA89 A0A0M9A0G5 A0A0A9XZF6 A0A1B6FRV0 A0A1B6CXD3 C4WSX4 A0A0P4VZJ7 A0A210PNU1 E9K6D5 T1HVL4 A0A0P6HW74 J9K6V6 E9H1B9 A0A0N8D1I6 A0A2R5LFE8 A0A2H8TJH4 A0A2T7NNE3 A0A0P6ASI4 A0A2S2R7S4 A0A1Q3FJB3 A0A131ZBC3 A0A224YL29 B0W1T5 G3MKX9 A0A023FLN2 A0A023FTP3 Q16G90 A0A2J7R5F5 C3YTG4 A0A023EF32 A0A3S3SMC1 U5EWY2 A0A2L2YA92 A0A3Q3KTS6 A0A3Q3KH25 L7LUW6 A0A087TSL1 A0A2L2YA94 A0A0L8G645 V5IIH3 T1JGY5 A0A3Q3KJ66 A0A131Y7F2 V5HVW0 A0A2A3EAY5 A0A3B4YL21 A0A3B4TWL8 A0A2U9BK68 W5J9F6 Q7SZ72 A0A182R7Y6 A0A182F3A2 A0A091NKG5 A0A131YBU8 A0A093I3Y3
Ontologies
PANTHER
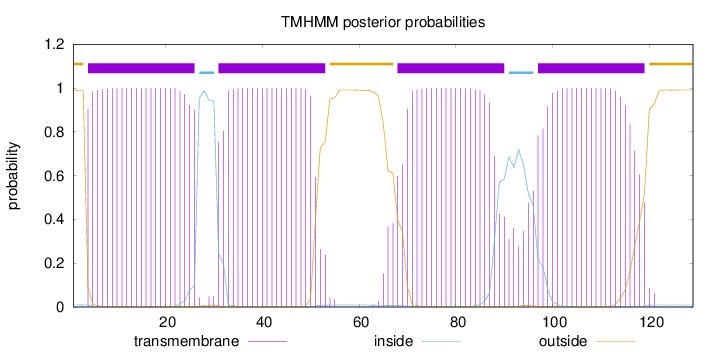
Topology
Length:
129
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
88.36934
Exp number, first 60 AAs:
43.4554
Total prob of N-in:
0.00964
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 26
inside
27 - 30
TMhelix
31 - 53
outside
54 - 67
TMhelix
68 - 90
inside
91 - 96
TMhelix
97 - 119
outside
120 - 129
Population Genetic Test Statistics
Pi
192.438062
Theta
2.693713
Tajima's D
0.317866
CLR
3.325517
CSRT
0.504974751262437
Interpretation
Uncertain
