Gene
KWMTBOMO11454
Pre Gene Modal
BGIBMGA001896
Annotation
PREDICTED:_uncharacterized_protein_LOC105389615_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.066
Sequence
CDS
ATGTACGAGCGACTACCGAATCAACAGTTACATGGATTTGACGATGACGTTGTACGAGAAACTGCACCGCCATTCCGCGTGTTGGAGAAATGCCAGGATCTATGTTTACGTGATAGATCTGGAAACAGCCTAGTCAGGACCTGCAACTCTATAGATTTTCAACCAGGGGCCCGTATAGCGGCCTTCAGTCCGGAACCAGAGTACGAGGAGTCAACATGCTACTTAACCAGAGAGCAGGCTACACCTGAAGGGATTGGGACTTTAATGATTGTCCCTAACAGTGTGCATTTTAATGAAATTTGTCTCACGTCAAATCGGCCTGAACGAGAATGTCCGTCGCGTCGCTATGTTTTTGAACGTCACGCACGGAAACGTTTGAAGCTACCGCCCTCAGACCTAAAGGAAATAATGGTCGCCAATCGCACCGAATGCGAGGACAAGTGCTTGGGAGAATTCAGCTTTGTCTGCCGTTCTGCGACTTATGATTCGGCTTTAAGAACATGCTCCCTCAGTAGATTTACGAGACGCACCCACCCGGAGTTATTGGAGGACGACCACAACGCTGATTACTTGGAAAATACATGTCTTAATGTGGCCTAA
Protein
MYERLPNQQLHGFDDDVVRETAPPFRVLEKCQDLCLRDRSGNSLVRTCNSIDFQPGARIAAFSPEPEYEESTCYLTREQATPEGIGTLMIVPNSVHFNEICLTSNRPERECPSRRYVFERHARKRLKLPPSDLKEIMVANRTECEDKCLGEFSFVCRSATYDSALRTCSLSRFTRRTHPELLEDDHNADYLENTCLNVA
Summary
Uniprot
A0A194QJF1
H9IXB3
A0A2A4JED9
A0A0N1PJM0
A0A2A4JFF2
A0A2A4JF45
+ More
A0A212EVI3 A0A2H1WV32 W8C174 A0A0K8WAG5 A0A1B6EYW8 Q1WWD1 A0A336LTK1 A0A182G6M4 A0A336LTL3 A0A1A9WP94 A0A1A9UV32 A0A1W4U5N1 A0A1A9YJ04 A0A0K8U2Z1 A0A1B0FN27 A0A1B0AAP1 A0A0B4LF37 A0A1B0BSL7 B4HNF3 A0A1W4UGL8 B4P7G4 B4MYB6 A0A1I8PQ32 A1Z8J3 Q32KF1 A0A1W4UGH8 B3NSI7 A0A0J9TYZ5 Q9BMD4 A0A1W4U500 B4KR75 B4QB78 B4J8W1 A1Z8J4 B4MEC9 Q9BMD5 B3MF92 A0A1I8MY63 A0A182MAX7 A0A3B0JFB9 B5E0A1 A0A3B0JKK1 A0A0J9RAB3 A0A2C9GRP2 A0A0J7NTZ9 B4GGH2 A0A1S4FK22 A0A182VE60 A0A0M4ECI2 A0A182WWW6 A0A1J1I6X9 A0A182TPY7 A0A0K8UYR3 Q7PTF3 A0A182KQB0 A0A0A1WII0 Q16Y69 A0A182SAF1 A0A182RF54 A0A0L0BPA5 A0A2C9GRZ8 A0A182YEI6 W5JK04 B0VZB8 A0A182Q5P2 A0A182FDG2 A0A2Y9D1G5 A0A195BWF1 A0A2Y9D1N7 A0A151JQX2 A0A182P472 A0A151IBX2 A0A158NCF5 A0A182JQG9 A0A195F9C1 A0A151WMX4 A0A084VZ46 A0A182WLF8 E2B676 A0A182J7H9 E2AYS0 A0A2S2N7W5 A0A154PP16 A0A087ZV55 D6WSE9 J9JPB7 A0A2A3E6U1 A0A026X1V9 A0A3L8DFM8 A0A2J7QD73 A0A2J7QD69 T1HW46 A0A2S2Q0X7 K7J454
A0A212EVI3 A0A2H1WV32 W8C174 A0A0K8WAG5 A0A1B6EYW8 Q1WWD1 A0A336LTK1 A0A182G6M4 A0A336LTL3 A0A1A9WP94 A0A1A9UV32 A0A1W4U5N1 A0A1A9YJ04 A0A0K8U2Z1 A0A1B0FN27 A0A1B0AAP1 A0A0B4LF37 A0A1B0BSL7 B4HNF3 A0A1W4UGL8 B4P7G4 B4MYB6 A0A1I8PQ32 A1Z8J3 Q32KF1 A0A1W4UGH8 B3NSI7 A0A0J9TYZ5 Q9BMD4 A0A1W4U500 B4KR75 B4QB78 B4J8W1 A1Z8J4 B4MEC9 Q9BMD5 B3MF92 A0A1I8MY63 A0A182MAX7 A0A3B0JFB9 B5E0A1 A0A3B0JKK1 A0A0J9RAB3 A0A2C9GRP2 A0A0J7NTZ9 B4GGH2 A0A1S4FK22 A0A182VE60 A0A0M4ECI2 A0A182WWW6 A0A1J1I6X9 A0A182TPY7 A0A0K8UYR3 Q7PTF3 A0A182KQB0 A0A0A1WII0 Q16Y69 A0A182SAF1 A0A182RF54 A0A0L0BPA5 A0A2C9GRZ8 A0A182YEI6 W5JK04 B0VZB8 A0A182Q5P2 A0A182FDG2 A0A2Y9D1G5 A0A195BWF1 A0A2Y9D1N7 A0A151JQX2 A0A182P472 A0A151IBX2 A0A158NCF5 A0A182JQG9 A0A195F9C1 A0A151WMX4 A0A084VZ46 A0A182WLF8 E2B676 A0A182J7H9 E2AYS0 A0A2S2N7W5 A0A154PP16 A0A087ZV55 D6WSE9 J9JPB7 A0A2A3E6U1 A0A026X1V9 A0A3L8DFM8 A0A2J7QD73 A0A2J7QD69 T1HW46 A0A2S2Q0X7 K7J454
Pubmed
26354079
19121390
22118469
24495485
26483478
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 17550304 26109357 26109356 22936249 11239432 25315136 15632085 17510324 12364791 14747013 17210077 20966253 25830018 26108605 25244985 20920257 23761445 21347285 24438588 20798317 18362917 19820115 24508170 30249741 20075255
12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 17550304 26109357 26109356 22936249 11239432 25315136 15632085 17510324 12364791 14747013 17210077 20966253 25830018 26108605 25244985 20920257 23761445 21347285 24438588 20798317 18362917 19820115 24508170 30249741 20075255
EMBL
KQ458575
KPJ05698.1
BABH01018024
BABH01018025
BABH01018026
NWSH01001679
+ More
PCG70437.1 KQ460044 KPJ18342.1 PCG70438.1 PCG70439.1 AGBW02012181 OWR45493.1 ODYU01011287 SOQ56929.1 GAMC01001113 JAC05443.1 GDHF01004136 JAI48178.1 GECZ01026560 JAS43209.1 BT024975 ABE01205.1 UFQS01000185 UFQT01000185 SSX00884.1 SSX21264.1 JXUM01045188 KQ561429 KXJ78612.1 SSX00885.1 SSX21265.1 GDHF01031604 JAI20710.1 CCAG010002874 AE013599 AHN56110.1 JXJN01019758 CH480816 EDW47389.1 CM000158 EDW89999.1 CH963894 EDW77105.2 AAM68728.2 AAS64916.1 BT023928 ABB36432.1 CH954179 EDV56489.2 CM002911 KMY92970.1 AF334032 AAK09434.1 CH933808 EDW08262.1 CM000362 EDX06611.1 CH916367 EDW02401.1 AAF58668.2 AAM68729.1 CH940662 EDW58894.1 AF334031 AAK09433.1 CH902619 EDV35566.2 AXCM01012009 OUUW01000001 SPP74000.1 CM000071 EDY69318.3 SPP73999.1 KMY92971.1 APCN01000155 LBMM01001683 KMQ95890.1 CH479183 EDW35592.1 CP012524 ALC41440.1 CVRI01000042 CRK95498.1 GDHF01020520 JAI31794.1 AAAB01008807 EAA04001.5 GBXI01015801 JAC98490.1 CH477523 EAT39570.1 JRES01001578 KNC21851.1 ADMH02001280 ETN63230.1 DS231813 EDS25694.1 AXCN02000424 AXCN02000425 KQ976396 KYM92912.1 KQ978619 KYN29766.1 KQ978076 KYM97227.1 ADTU01011868 ADTU01011869 ADTU01011870 KQ981727 KYN37038.1 KQ982934 KYQ49170.1 ATLV01018601 KE525239 KFB43240.1 GL445930 EFN88769.1 GL443983 EFN61433.1 GGMR01000626 MBY13245.1 KQ435007 KZC13609.1 KQ971352 EFA06632.2 ABLF02040773 KZ288362 PBC26946.1 KK107034 EZA62053.1 QOIP01000008 RLU19270.1 NEVH01015827 PNF26529.1 PNF26531.1 ACPB03001675 GGMS01002118 MBY71321.1 AAZX01007897
PCG70437.1 KQ460044 KPJ18342.1 PCG70438.1 PCG70439.1 AGBW02012181 OWR45493.1 ODYU01011287 SOQ56929.1 GAMC01001113 JAC05443.1 GDHF01004136 JAI48178.1 GECZ01026560 JAS43209.1 BT024975 ABE01205.1 UFQS01000185 UFQT01000185 SSX00884.1 SSX21264.1 JXUM01045188 KQ561429 KXJ78612.1 SSX00885.1 SSX21265.1 GDHF01031604 JAI20710.1 CCAG010002874 AE013599 AHN56110.1 JXJN01019758 CH480816 EDW47389.1 CM000158 EDW89999.1 CH963894 EDW77105.2 AAM68728.2 AAS64916.1 BT023928 ABB36432.1 CH954179 EDV56489.2 CM002911 KMY92970.1 AF334032 AAK09434.1 CH933808 EDW08262.1 CM000362 EDX06611.1 CH916367 EDW02401.1 AAF58668.2 AAM68729.1 CH940662 EDW58894.1 AF334031 AAK09433.1 CH902619 EDV35566.2 AXCM01012009 OUUW01000001 SPP74000.1 CM000071 EDY69318.3 SPP73999.1 KMY92971.1 APCN01000155 LBMM01001683 KMQ95890.1 CH479183 EDW35592.1 CP012524 ALC41440.1 CVRI01000042 CRK95498.1 GDHF01020520 JAI31794.1 AAAB01008807 EAA04001.5 GBXI01015801 JAC98490.1 CH477523 EAT39570.1 JRES01001578 KNC21851.1 ADMH02001280 ETN63230.1 DS231813 EDS25694.1 AXCN02000424 AXCN02000425 KQ976396 KYM92912.1 KQ978619 KYN29766.1 KQ978076 KYM97227.1 ADTU01011868 ADTU01011869 ADTU01011870 KQ981727 KYN37038.1 KQ982934 KYQ49170.1 ATLV01018601 KE525239 KFB43240.1 GL445930 EFN88769.1 GL443983 EFN61433.1 GGMR01000626 MBY13245.1 KQ435007 KZC13609.1 KQ971352 EFA06632.2 ABLF02040773 KZ288362 PBC26946.1 KK107034 EZA62053.1 QOIP01000008 RLU19270.1 NEVH01015827 PNF26529.1 PNF26531.1 ACPB03001675 GGMS01002118 MBY71321.1 AAZX01007897
Proteomes
UP000053268
UP000005204
UP000218220
UP000053240
UP000007151
UP000069940
+ More
UP000249989 UP000091820 UP000078200 UP000192221 UP000092443 UP000092444 UP000092445 UP000000803 UP000092460 UP000001292 UP000002282 UP000007798 UP000095300 UP000008711 UP000009192 UP000000304 UP000001070 UP000008792 UP000007801 UP000095301 UP000075883 UP000268350 UP000001819 UP000075840 UP000036403 UP000008744 UP000075903 UP000092553 UP000076407 UP000183832 UP000075902 UP000007062 UP000075882 UP000008820 UP000075901 UP000075900 UP000037069 UP000076408 UP000000673 UP000002320 UP000075886 UP000069272 UP000075884 UP000078540 UP000078492 UP000075885 UP000078542 UP000005205 UP000075881 UP000078541 UP000075809 UP000030765 UP000075920 UP000008237 UP000075880 UP000000311 UP000076502 UP000005203 UP000007266 UP000007819 UP000242457 UP000053097 UP000279307 UP000235965 UP000015103 UP000002358
UP000249989 UP000091820 UP000078200 UP000192221 UP000092443 UP000092444 UP000092445 UP000000803 UP000092460 UP000001292 UP000002282 UP000007798 UP000095300 UP000008711 UP000009192 UP000000304 UP000001070 UP000008792 UP000007801 UP000095301 UP000075883 UP000268350 UP000001819 UP000075840 UP000036403 UP000008744 UP000075903 UP000092553 UP000076407 UP000183832 UP000075902 UP000007062 UP000075882 UP000008820 UP000075901 UP000075900 UP000037069 UP000076408 UP000000673 UP000002320 UP000075886 UP000069272 UP000075884 UP000078540 UP000078492 UP000075885 UP000078542 UP000005205 UP000075881 UP000078541 UP000075809 UP000030765 UP000075920 UP000008237 UP000075880 UP000000311 UP000076502 UP000005203 UP000007266 UP000007819 UP000242457 UP000053097 UP000279307 UP000235965 UP000015103 UP000002358
Interpro
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
A0A194QJF1
H9IXB3
A0A2A4JED9
A0A0N1PJM0
A0A2A4JFF2
A0A2A4JF45
+ More
A0A212EVI3 A0A2H1WV32 W8C174 A0A0K8WAG5 A0A1B6EYW8 Q1WWD1 A0A336LTK1 A0A182G6M4 A0A336LTL3 A0A1A9WP94 A0A1A9UV32 A0A1W4U5N1 A0A1A9YJ04 A0A0K8U2Z1 A0A1B0FN27 A0A1B0AAP1 A0A0B4LF37 A0A1B0BSL7 B4HNF3 A0A1W4UGL8 B4P7G4 B4MYB6 A0A1I8PQ32 A1Z8J3 Q32KF1 A0A1W4UGH8 B3NSI7 A0A0J9TYZ5 Q9BMD4 A0A1W4U500 B4KR75 B4QB78 B4J8W1 A1Z8J4 B4MEC9 Q9BMD5 B3MF92 A0A1I8MY63 A0A182MAX7 A0A3B0JFB9 B5E0A1 A0A3B0JKK1 A0A0J9RAB3 A0A2C9GRP2 A0A0J7NTZ9 B4GGH2 A0A1S4FK22 A0A182VE60 A0A0M4ECI2 A0A182WWW6 A0A1J1I6X9 A0A182TPY7 A0A0K8UYR3 Q7PTF3 A0A182KQB0 A0A0A1WII0 Q16Y69 A0A182SAF1 A0A182RF54 A0A0L0BPA5 A0A2C9GRZ8 A0A182YEI6 W5JK04 B0VZB8 A0A182Q5P2 A0A182FDG2 A0A2Y9D1G5 A0A195BWF1 A0A2Y9D1N7 A0A151JQX2 A0A182P472 A0A151IBX2 A0A158NCF5 A0A182JQG9 A0A195F9C1 A0A151WMX4 A0A084VZ46 A0A182WLF8 E2B676 A0A182J7H9 E2AYS0 A0A2S2N7W5 A0A154PP16 A0A087ZV55 D6WSE9 J9JPB7 A0A2A3E6U1 A0A026X1V9 A0A3L8DFM8 A0A2J7QD73 A0A2J7QD69 T1HW46 A0A2S2Q0X7 K7J454
A0A212EVI3 A0A2H1WV32 W8C174 A0A0K8WAG5 A0A1B6EYW8 Q1WWD1 A0A336LTK1 A0A182G6M4 A0A336LTL3 A0A1A9WP94 A0A1A9UV32 A0A1W4U5N1 A0A1A9YJ04 A0A0K8U2Z1 A0A1B0FN27 A0A1B0AAP1 A0A0B4LF37 A0A1B0BSL7 B4HNF3 A0A1W4UGL8 B4P7G4 B4MYB6 A0A1I8PQ32 A1Z8J3 Q32KF1 A0A1W4UGH8 B3NSI7 A0A0J9TYZ5 Q9BMD4 A0A1W4U500 B4KR75 B4QB78 B4J8W1 A1Z8J4 B4MEC9 Q9BMD5 B3MF92 A0A1I8MY63 A0A182MAX7 A0A3B0JFB9 B5E0A1 A0A3B0JKK1 A0A0J9RAB3 A0A2C9GRP2 A0A0J7NTZ9 B4GGH2 A0A1S4FK22 A0A182VE60 A0A0M4ECI2 A0A182WWW6 A0A1J1I6X9 A0A182TPY7 A0A0K8UYR3 Q7PTF3 A0A182KQB0 A0A0A1WII0 Q16Y69 A0A182SAF1 A0A182RF54 A0A0L0BPA5 A0A2C9GRZ8 A0A182YEI6 W5JK04 B0VZB8 A0A182Q5P2 A0A182FDG2 A0A2Y9D1G5 A0A195BWF1 A0A2Y9D1N7 A0A151JQX2 A0A182P472 A0A151IBX2 A0A158NCF5 A0A182JQG9 A0A195F9C1 A0A151WMX4 A0A084VZ46 A0A182WLF8 E2B676 A0A182J7H9 E2AYS0 A0A2S2N7W5 A0A154PP16 A0A087ZV55 D6WSE9 J9JPB7 A0A2A3E6U1 A0A026X1V9 A0A3L8DFM8 A0A2J7QD73 A0A2J7QD69 T1HW46 A0A2S2Q0X7 K7J454
Ontologies
GO
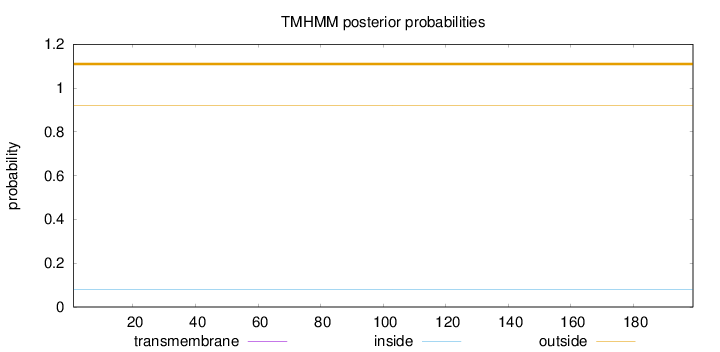
Topology
Length:
199
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00042
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07875
outside
1 - 199
Population Genetic Test Statistics
Pi
148.755278
Theta
56.82222
Tajima's D
1.978732
CLR
0.254936
CSRT
0.88285585720714
Interpretation
Uncertain
