Pre Gene Modal
BGIBMGA001927
Annotation
farnesyl_diphosphate_synthase_2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.716 Mitochondrial Reliability : 1.197
Sequence
CDS
ATGGCCAGCGCCATACAAACTTTATGGCGCACTTCAATGGGTCAACATTTAGATCATGTAACAGGAAATCGAAAAACAGACTATTCTTCATTTACTCTGGATCGTTACTACACTATTATAAAATACAAGGCCGCATATTACACCTACAATTTACCCGTTTCCCTTGGTTTACTACTAGCTGAAAATGTGGATGAAAAAATTTATAAATCTGCTCAAGACATTTGCTTGGAAATAGGAACGATGTTCCAAATTCAGGACGATTTCATAGACTGCTTCGGTGATGAAATAAAAACTGGTAAAGTTGGTACAGATATACAAGAGCGCAAATGTACTTGGTTGGCAGTTCAAGCGCTGCAGCGCTGTACTGAAGCACAACGTACAGTATTCAAAGCCTGTTATGGTAGCAGTGAACCAGCACATGTGGAACGCATAAAACGACTTTACGAAGACCTTCACCTCCCGCAGATATACAAGCATCAAGAAAAAGCGATGTACGACAATATTATAAGACAAATTGAAAATATACCAATAGAGGCCGCCCGCGTTCTCTTTAAAAAGTTGTTGGATATTACGTACAATAGGCAACATTAA
Protein
MASAIQTLWRTSMGQHLDHVTGNRKTDYSSFTLDRYYTIIKYKAAYYTYNLPVSLGLLLAENVDEKIYKSAQDICLEIGTMFQIQDDFIDCFGDEIKTGKVGTDIQERKCTWLAVQALQRCTEAQRTVFKACYGSSEPAHVERIKRLYEDLHLPQIYKHQEKAMYDNIIRQIENIPIEAARVLFKKLLDITYNRQH
Summary
Similarity
Belongs to the FPP/GGPP synthase family.
Uniprot
A5A7A5
H9IXE4
A0A0N1PIW0
A0A194QQ94
A0A212EVK2
Q1X8X9
+ More
Q1XAB1 A0A2H1VJC4 S4P0U8 A0A2A4IUH1 A0A2H1WY54 A0A2A4JEQ9 A0A0L7L6A6 A5A7A6 A0A1W5ZM49 A0A0L7K4G9 A0A2A4JF30 A0A2A4JGC1 A0A2A4JFI5 A0A140DKW6 A0A1B6EYU0 A0A1B6E8U3 A0A2P8ZM10 A0A0C9R8J4 A0A182TY89 A0A2H1WV31 Q1XAB0 Q1X8X8 A0A182MIA4 S4PI76 E9J201 A0A1B0DI83 A0A182IT14 A0A182WWX1 A0A182NK80 A0A182RF58 U5ETY6 A0A182WLG4 A0A182QJE2 A0A182JX98 A0A182V441 A0A182I2G9 Q7QIW5 A0A182P468 A0A0N1IG58 A0A024G1U7 A0A182KQC2 A0A024G144 A0A182YEJ2 A0A2J7PU44 A0A024G0B1 A0A067RTI2 A0A194QJJ2 A0A0N1PFP8 A0A2A4JG86 A0A1W5ZM48 A0A2M4BNI6 A0A2M3Z093 A0A2M4A321 A0A182H5G9 A0A2M4CPQ8 A0A2M4CPT7 A0A1L8DZJ9 A0A1L8DZI7 A0A1Q3FYU3 O76820 W5J8W7 Q17F78 A0A023ES27 K3WEV8 A0A1B0CJ89 K7IWR9 Q1XAA9 A0A1Z5JVE3 A0A1Y1LLN4 A0A1L3KPV5 Q56CY7 I1VJ84 A0A0L7QWI9 A0A232EKP8 A0A0K2TWZ6 E1Z2N1 A0A2H1VL04 A0A024GRX7 A0A1Z5K194 A0A182FGW5 A0A0N9EI79 A0A0P4VIE8 T1ID56 A5A7A4 Q95P28 H9IXE3 A0A0N1IP44 A0A0G4EIF1 B0VZA8 A0A2P6U1R9 A0A2P6VM23 A0A1W7R8C9 A0A212EVH6 A0A2H1WST5
Q1XAB1 A0A2H1VJC4 S4P0U8 A0A2A4IUH1 A0A2H1WY54 A0A2A4JEQ9 A0A0L7L6A6 A5A7A6 A0A1W5ZM49 A0A0L7K4G9 A0A2A4JF30 A0A2A4JGC1 A0A2A4JFI5 A0A140DKW6 A0A1B6EYU0 A0A1B6E8U3 A0A2P8ZM10 A0A0C9R8J4 A0A182TY89 A0A2H1WV31 Q1XAB0 Q1X8X8 A0A182MIA4 S4PI76 E9J201 A0A1B0DI83 A0A182IT14 A0A182WWX1 A0A182NK80 A0A182RF58 U5ETY6 A0A182WLG4 A0A182QJE2 A0A182JX98 A0A182V441 A0A182I2G9 Q7QIW5 A0A182P468 A0A0N1IG58 A0A024G1U7 A0A182KQC2 A0A024G144 A0A182YEJ2 A0A2J7PU44 A0A024G0B1 A0A067RTI2 A0A194QJJ2 A0A0N1PFP8 A0A2A4JG86 A0A1W5ZM48 A0A2M4BNI6 A0A2M3Z093 A0A2M4A321 A0A182H5G9 A0A2M4CPQ8 A0A2M4CPT7 A0A1L8DZJ9 A0A1L8DZI7 A0A1Q3FYU3 O76820 W5J8W7 Q17F78 A0A023ES27 K3WEV8 A0A1B0CJ89 K7IWR9 Q1XAA9 A0A1Z5JVE3 A0A1Y1LLN4 A0A1L3KPV5 Q56CY7 I1VJ84 A0A0L7QWI9 A0A232EKP8 A0A0K2TWZ6 E1Z2N1 A0A2H1VL04 A0A024GRX7 A0A1Z5K194 A0A182FGW5 A0A0N9EI79 A0A0P4VIE8 T1ID56 A5A7A4 Q95P28 H9IXE3 A0A0N1IP44 A0A0G4EIF1 B0VZA8 A0A2P6U1R9 A0A2P6VM23 A0A1W7R8C9 A0A212EVH6 A0A2H1WST5
Pubmed
17628279
19121390
26354079
22118469
29183817
23622113
+ More
26227816 28382786 29403074 21282665 12364791 14747013 17210077 20966253 25244985 24845553 26483478 10336620 20920257 23761445 17510324 24945155 20075255 25634988 28004739 15983375 22516182 23537049 24167290 28648823 20852019 27129103 29178410
26227816 28382786 29403074 21282665 12364791 14747013 17210077 20966253 25244985 24845553 26483478 10336620 20920257 23761445 17510324 24945155 20075255 25634988 28004739 15983375 22516182 23537049 24167290 28648823 20852019 27129103 29178410
EMBL
AB274996
BAF62114.1
BABH01018027
BABH01018028
BABH01018029
KQ460044
+ More
KPJ18340.1 KQ458575 KPJ05696.1 AGBW02012181 OWR45491.1 AY962307 AAY26574.1 AY954919 AAY33485.1 ODYU01002886 SOQ40935.1 GAIX01009201 JAA83359.1 NWSH01006480 PCG63405.1 ODYU01011963 SOQ58005.1 NWSH01001679 PCG70441.1 JTDY01002731 KOB70829.1 AB274997 BAF62115.1 KY421779 ARI71315.1 JTDY01011123 KOB57504.1 NWSH01001773 PCG70184.1 PCG70442.1 PCG70183.1 KT748800 AMK48581.1 GECZ01026652 JAS43117.1 GEDC01002947 JAS34351.1 PYGN01000019 PSN57541.1 GBYB01003116 JAG72883.1 ODYU01011280 SOQ56918.1 AY954920 AAY33486.1 AH014859 AY962308 AAY26575.1 AXCM01017175 GAIX01005505 JAA87055.1 GL767700 EFZ13152.1 AJVK01015478 GANO01002569 JAB57302.1 AXCN02000425 APCN01000156 AAAB01008807 EAA04004.4 KPJ18341.1 CAIX01000007 CCI40283.1 CCI40281.1 NEVH01021208 PNF19862.1 CCI40282.1 KK852473 KDR23124.1 KPJ05697.1 KQ460772 KPJ12445.1 PCG70440.1 KY421778 ARI71314.1 GGFJ01005474 MBW54615.1 GGFM01001150 MBW21901.1 GGFK01001820 MBW35141.1 JXUM01026170 KQ560746 KXJ81031.1 GGFL01003136 MBW67314.1 GGFL01003135 MBW67313.1 GFDF01002193 JAV11891.1 GFDF01002194 JAV11890.1 GFDL01002296 JAV32749.1 AJ009962 CAA08918.2 ADMH02001942 ETN60396.1 CH477273 EAT45207.1 GAPW01001563 JAC12035.1 GL376603 AJWK01014316 AAZX01000604 AY954921 AAY33487.1 BDSP01000123 GAX17802.1 GEZM01054111 JAV73841.1 KU163588 LC176696 APG79413.1 BBA21113.1 AY966009 AAX78435.1 APGK01054108 APGK01054109 JQ855705 KB741247 KB632281 AFI45068.1 ENN72022.1 ERL91359.1 KQ414716 KOC62899.1 NNAY01003758 OXU18892.1 HACA01012535 CDW29896.1 GL433835 EFN59689.1 SOQ40934.1 CAIX01000307 CCI49531.1 BDSP01000142 GAX20074.1 KR140284 ALF44684.1 GDKW01002946 JAI53649.1 ACPB03002853 AB274995 BAF62113.1 AB072589 BAB69490.1 BABH01018026 KPJ12446.1 CDMY01000238 CEL95761.1 DS231813 EDS25700.1 LHPG02000002 PRW60267.1 LHPF02000003 PSC75133.1 GEHC01000234 JAV47411.1 OWR45492.1 ODYU01010797 SOQ56123.1
KPJ18340.1 KQ458575 KPJ05696.1 AGBW02012181 OWR45491.1 AY962307 AAY26574.1 AY954919 AAY33485.1 ODYU01002886 SOQ40935.1 GAIX01009201 JAA83359.1 NWSH01006480 PCG63405.1 ODYU01011963 SOQ58005.1 NWSH01001679 PCG70441.1 JTDY01002731 KOB70829.1 AB274997 BAF62115.1 KY421779 ARI71315.1 JTDY01011123 KOB57504.1 NWSH01001773 PCG70184.1 PCG70442.1 PCG70183.1 KT748800 AMK48581.1 GECZ01026652 JAS43117.1 GEDC01002947 JAS34351.1 PYGN01000019 PSN57541.1 GBYB01003116 JAG72883.1 ODYU01011280 SOQ56918.1 AY954920 AAY33486.1 AH014859 AY962308 AAY26575.1 AXCM01017175 GAIX01005505 JAA87055.1 GL767700 EFZ13152.1 AJVK01015478 GANO01002569 JAB57302.1 AXCN02000425 APCN01000156 AAAB01008807 EAA04004.4 KPJ18341.1 CAIX01000007 CCI40283.1 CCI40281.1 NEVH01021208 PNF19862.1 CCI40282.1 KK852473 KDR23124.1 KPJ05697.1 KQ460772 KPJ12445.1 PCG70440.1 KY421778 ARI71314.1 GGFJ01005474 MBW54615.1 GGFM01001150 MBW21901.1 GGFK01001820 MBW35141.1 JXUM01026170 KQ560746 KXJ81031.1 GGFL01003136 MBW67314.1 GGFL01003135 MBW67313.1 GFDF01002193 JAV11891.1 GFDF01002194 JAV11890.1 GFDL01002296 JAV32749.1 AJ009962 CAA08918.2 ADMH02001942 ETN60396.1 CH477273 EAT45207.1 GAPW01001563 JAC12035.1 GL376603 AJWK01014316 AAZX01000604 AY954921 AAY33487.1 BDSP01000123 GAX17802.1 GEZM01054111 JAV73841.1 KU163588 LC176696 APG79413.1 BBA21113.1 AY966009 AAX78435.1 APGK01054108 APGK01054109 JQ855705 KB741247 KB632281 AFI45068.1 ENN72022.1 ERL91359.1 KQ414716 KOC62899.1 NNAY01003758 OXU18892.1 HACA01012535 CDW29896.1 GL433835 EFN59689.1 SOQ40934.1 CAIX01000307 CCI49531.1 BDSP01000142 GAX20074.1 KR140284 ALF44684.1 GDKW01002946 JAI53649.1 ACPB03002853 AB274995 BAF62113.1 AB072589 BAB69490.1 BABH01018026 KPJ12446.1 CDMY01000238 CEL95761.1 DS231813 EDS25700.1 LHPG02000002 PRW60267.1 LHPF02000003 PSC75133.1 GEHC01000234 JAV47411.1 OWR45492.1 ODYU01010797 SOQ56123.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000037510
+ More
UP000245037 UP000075902 UP000075883 UP000092462 UP000075880 UP000076407 UP000075884 UP000075900 UP000075920 UP000075886 UP000075881 UP000075903 UP000075840 UP000007062 UP000075885 UP000053237 UP000075882 UP000076408 UP000235965 UP000027135 UP000069940 UP000249989 UP000000673 UP000008820 UP000092461 UP000002358 UP000019118 UP000030742 UP000053825 UP000215335 UP000008141 UP000069272 UP000015103 UP000041254 UP000002320 UP000239899 UP000239649
UP000245037 UP000075902 UP000075883 UP000092462 UP000075880 UP000076407 UP000075884 UP000075900 UP000075920 UP000075886 UP000075881 UP000075903 UP000075840 UP000007062 UP000075885 UP000053237 UP000075882 UP000076408 UP000235965 UP000027135 UP000069940 UP000249989 UP000000673 UP000008820 UP000092461 UP000002358 UP000019118 UP000030742 UP000053825 UP000215335 UP000008141 UP000069272 UP000015103 UP000041254 UP000002320 UP000239899 UP000239649
Interpro
Gene 3D
ProteinModelPortal
A5A7A5
H9IXE4
A0A0N1PIW0
A0A194QQ94
A0A212EVK2
Q1X8X9
+ More
Q1XAB1 A0A2H1VJC4 S4P0U8 A0A2A4IUH1 A0A2H1WY54 A0A2A4JEQ9 A0A0L7L6A6 A5A7A6 A0A1W5ZM49 A0A0L7K4G9 A0A2A4JF30 A0A2A4JGC1 A0A2A4JFI5 A0A140DKW6 A0A1B6EYU0 A0A1B6E8U3 A0A2P8ZM10 A0A0C9R8J4 A0A182TY89 A0A2H1WV31 Q1XAB0 Q1X8X8 A0A182MIA4 S4PI76 E9J201 A0A1B0DI83 A0A182IT14 A0A182WWX1 A0A182NK80 A0A182RF58 U5ETY6 A0A182WLG4 A0A182QJE2 A0A182JX98 A0A182V441 A0A182I2G9 Q7QIW5 A0A182P468 A0A0N1IG58 A0A024G1U7 A0A182KQC2 A0A024G144 A0A182YEJ2 A0A2J7PU44 A0A024G0B1 A0A067RTI2 A0A194QJJ2 A0A0N1PFP8 A0A2A4JG86 A0A1W5ZM48 A0A2M4BNI6 A0A2M3Z093 A0A2M4A321 A0A182H5G9 A0A2M4CPQ8 A0A2M4CPT7 A0A1L8DZJ9 A0A1L8DZI7 A0A1Q3FYU3 O76820 W5J8W7 Q17F78 A0A023ES27 K3WEV8 A0A1B0CJ89 K7IWR9 Q1XAA9 A0A1Z5JVE3 A0A1Y1LLN4 A0A1L3KPV5 Q56CY7 I1VJ84 A0A0L7QWI9 A0A232EKP8 A0A0K2TWZ6 E1Z2N1 A0A2H1VL04 A0A024GRX7 A0A1Z5K194 A0A182FGW5 A0A0N9EI79 A0A0P4VIE8 T1ID56 A5A7A4 Q95P28 H9IXE3 A0A0N1IP44 A0A0G4EIF1 B0VZA8 A0A2P6U1R9 A0A2P6VM23 A0A1W7R8C9 A0A212EVH6 A0A2H1WST5
Q1XAB1 A0A2H1VJC4 S4P0U8 A0A2A4IUH1 A0A2H1WY54 A0A2A4JEQ9 A0A0L7L6A6 A5A7A6 A0A1W5ZM49 A0A0L7K4G9 A0A2A4JF30 A0A2A4JGC1 A0A2A4JFI5 A0A140DKW6 A0A1B6EYU0 A0A1B6E8U3 A0A2P8ZM10 A0A0C9R8J4 A0A182TY89 A0A2H1WV31 Q1XAB0 Q1X8X8 A0A182MIA4 S4PI76 E9J201 A0A1B0DI83 A0A182IT14 A0A182WWX1 A0A182NK80 A0A182RF58 U5ETY6 A0A182WLG4 A0A182QJE2 A0A182JX98 A0A182V441 A0A182I2G9 Q7QIW5 A0A182P468 A0A0N1IG58 A0A024G1U7 A0A182KQC2 A0A024G144 A0A182YEJ2 A0A2J7PU44 A0A024G0B1 A0A067RTI2 A0A194QJJ2 A0A0N1PFP8 A0A2A4JG86 A0A1W5ZM48 A0A2M4BNI6 A0A2M3Z093 A0A2M4A321 A0A182H5G9 A0A2M4CPQ8 A0A2M4CPT7 A0A1L8DZJ9 A0A1L8DZI7 A0A1Q3FYU3 O76820 W5J8W7 Q17F78 A0A023ES27 K3WEV8 A0A1B0CJ89 K7IWR9 Q1XAA9 A0A1Z5JVE3 A0A1Y1LLN4 A0A1L3KPV5 Q56CY7 I1VJ84 A0A0L7QWI9 A0A232EKP8 A0A0K2TWZ6 E1Z2N1 A0A2H1VL04 A0A024GRX7 A0A1Z5K194 A0A182FGW5 A0A0N9EI79 A0A0P4VIE8 T1ID56 A5A7A4 Q95P28 H9IXE3 A0A0N1IP44 A0A0G4EIF1 B0VZA8 A0A2P6U1R9 A0A2P6VM23 A0A1W7R8C9 A0A212EVH6 A0A2H1WST5
PDB
6B07
E-value=1.88407e-54,
Score=533
Ontologies
GO
PANTHER
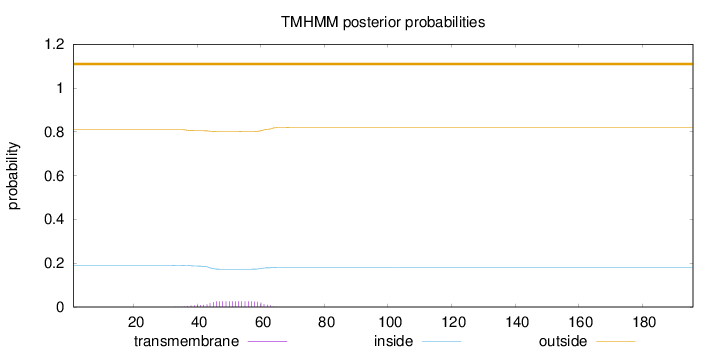
Topology
Length:
196
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.505279999999999
Exp number, first 60 AAs:
0.47302
Total prob of N-in:
0.19023
outside
1 - 196
Population Genetic Test Statistics
Pi
152.417917
Theta
139.312368
Tajima's D
0.258619
CLR
0.943263
CSRT
0.44252787360632
Interpretation
Uncertain
