Gene
KWMTBOMO11447
Pre Gene Modal
BGIBMGA001894
Annotation
PREDICTED:_uncharacterized_protein_LOC106132668_isoform_X2_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.525 PlasmaMembrane Reliability : 1.762
Sequence
CDS
ATGTCTAGCTTACGTTTAGATTCGTGCTGGGAACAAGAGGAAGACGGTAACACAAGTCGTCATGTAACTGGCGATCCAATAGCCTTGATATATAATGAAGGCCTTTGGAAAATAAACAAAGTATCTCCCTTGTACAATTTGCAATACAATGGTATTAAGTTGAAGCAGTATGGCTCTAAAATCCTTCAGGCACTCGTCAGCTCATTGCAAATGAGTAATACAGCAAAATACACTGTAGTACTTGAAGAACTACCACTACTGAAATACTCCGAAGAAGATTCTAGTGGAATACTAATAACAGTTAGATCATCACAAGAGGACAGCAATACTAAAAAAGTGGCATATGTTGCTATATTATTGTCTTGGGGTATAAGCATTAGTATGGAAAATGCAATACATCTACCTTATATGCTAGAACGCGGTGAACAGAGAGTTGGAACAGTTGTAAAAACAATTTTGCAGACTGCTTTTGATTGCAACATTAAACAGTTTAGTTTTACTCAACACCAACTTTTGAACTTTGGTTTCAGTTTTATAGACAGTGATACATCCCGGTCAAATGATCAATTTACACTTGTGTATAGAAGTCCACAGACTGAAGTCAAAAGTAAATTAAGTCTATCATTTGACATAGGAGATGTGAGACTGATTTGGAATGGAATAAAAGATGAAACAACAAAGAAAGCAGACTTAATAATCCTGGCATACCAGATATTACAAAATCAAATATTCAGTTTGGGTTACCTGGATGTTACTGTATTCGATTTGAGTGAAATTAATTTAACAAAAGCAGAAATAAAATGCAATGGAGGTGTGAAAATGAAAACGCCAGAAATTGTGAACTGCGTATTCACTGTTTTAAATGAAATTTGCTGTGATTTATGGCAGGATTTGAACATTACAGAGTGA
Protein
MSSLRLDSCWEQEEDGNTSRHVTGDPIALIYNEGLWKINKVSPLYNLQYNGIKLKQYGSKILQALVSSLQMSNTAKYTVVLEELPLLKYSEEDSSGILITVRSSQEDSNTKKVAYVAILLSWGISISMENAIHLPYMLERGEQRVGTVVKTILQTAFDCNIKQFSFTQHQLLNFGFSFIDSDTSRSNDQFTLVYRSPQTEVKSKLSLSFDIGDVRLIWNGIKDETTKKADLIILAYQILQNQIFSLGYLDVTVFDLSEINLTKAEIKCNGGVKMKTPEIVNCVFTVLNEICCDLWQDLNITE
Summary
Uniprot
H9IXB1
A0A2A4K363
A0A2H1W3H5
A0A212EVG4
A0A194QJI5
A0A0N1I8H0
+ More
S4NNI8 A0A0L7KHB3 A0A2J7RKN5 A0A067QX65 A0A0C9Q978 A0A1S3K664 A0A2R2MSD2 A0A1S3H1I0 A0A1S3GZN3 A0A088AJ36 A0A026W870 A0A3L8DYN7 A0A195BQI1 A0A158NDS1 A0A195EZF6 A0A195CM69 A0A2A3E6G7 A0A151X4P6 K7ISD0 E9J519 A0A195D7M7 A0A0M8ZW33 A0A310SEQ0 C1C4S8 A0A232F773 A0A091JNI2 A0A094LG46 A0A3S2MGF1 A0A1B6CL68 A0A2I0MBS0 A0A093JF37 A0A1V4JXH2 A0A0P5BF54 A0A0P5EF24 A0A1B6L7Y6 A0A091VZG8 A0A154PG61 A0A0N8AHX3 A0A3P9L7G8 A0A0P5X9N8 A0A0P5WM11 A0A091JYJ4 A0A0P7WU96 A0A1W5A7N1 A0A093BKM9 A0A3B5KA17 A0A093R1B7 A0A087VQM7 A0A3P9JHC3 A0A087QVK6 A0A091WI06 A0A0A0B049 A0A1A8DTH9 A0A091RD97 A0A1S3R182 A0A091N7S0 A0A093Q463 H2LJ76 A0A1A8L523 A0A091TRF0 A0A1A8UZ18 H3AUJ6 A0A2I0UHS7 A0A1A7ZYB5 A0A091T855 A0A0Q3U511 A0A060VVV0 A0A060WN69 A0A093PPY7 H2TXD4 A0A0P5RAL4 A0A0N8DLC7 G1KR38 A0A1A8J6B7 A0A3B3BHQ5 A0A3B3Y555 V9KYT9 A0A091SAU5 A0A087X579 A0A3P8Y0F4 A0A091FY01 A0A093IP24 W5MS61 A0A093CQC8 A0A3B4H300 A0A3B5QRX3
S4NNI8 A0A0L7KHB3 A0A2J7RKN5 A0A067QX65 A0A0C9Q978 A0A1S3K664 A0A2R2MSD2 A0A1S3H1I0 A0A1S3GZN3 A0A088AJ36 A0A026W870 A0A3L8DYN7 A0A195BQI1 A0A158NDS1 A0A195EZF6 A0A195CM69 A0A2A3E6G7 A0A151X4P6 K7ISD0 E9J519 A0A195D7M7 A0A0M8ZW33 A0A310SEQ0 C1C4S8 A0A232F773 A0A091JNI2 A0A094LG46 A0A3S2MGF1 A0A1B6CL68 A0A2I0MBS0 A0A093JF37 A0A1V4JXH2 A0A0P5BF54 A0A0P5EF24 A0A1B6L7Y6 A0A091VZG8 A0A154PG61 A0A0N8AHX3 A0A3P9L7G8 A0A0P5X9N8 A0A0P5WM11 A0A091JYJ4 A0A0P7WU96 A0A1W5A7N1 A0A093BKM9 A0A3B5KA17 A0A093R1B7 A0A087VQM7 A0A3P9JHC3 A0A087QVK6 A0A091WI06 A0A0A0B049 A0A1A8DTH9 A0A091RD97 A0A1S3R182 A0A091N7S0 A0A093Q463 H2LJ76 A0A1A8L523 A0A091TRF0 A0A1A8UZ18 H3AUJ6 A0A2I0UHS7 A0A1A7ZYB5 A0A091T855 A0A0Q3U511 A0A060VVV0 A0A060WN69 A0A093PPY7 H2TXD4 A0A0P5RAL4 A0A0N8DLC7 G1KR38 A0A1A8J6B7 A0A3B3BHQ5 A0A3B3Y555 V9KYT9 A0A091SAU5 A0A087X579 A0A3P8Y0F4 A0A091FY01 A0A093IP24 W5MS61 A0A093CQC8 A0A3B4H300 A0A3B5QRX3
Pubmed
EMBL
BABH01018031
AK289344
BAM73871.1
NWSH01000239
PCG78092.1
ODYU01006096
+ More
SOQ47640.1 AGBW02012181 OWR45488.1 KQ458575 KPJ05687.1 KQ460772 KPJ12449.1 GAIX01013911 JAA78649.1 JTDY01009783 KOB62713.1 NEVH01002717 PNF41403.1 KK853253 KDR09309.1 GBYB01010908 GBYB01010909 JAG80675.1 JAG80676.1 KK107371 EZA51846.1 QOIP01000002 RLU25333.1 KQ976424 KYM88393.1 ADTU01012852 KQ981906 KYN33259.1 KQ977600 KYN01567.1 KZ288368 PBC26816.1 KQ982548 KYQ55198.1 GL768123 EFZ12079.1 KQ981153 KYN08890.1 KQ435851 KOX70729.1 KQ760629 OAD59737.1 BT081857 ACO51988.1 NNAY01000827 OXU26299.1 KK502178 KFP21325.1 KL357092 KFZ62944.1 CM012438 RVE75381.1 GEDC01030128 GEDC01030064 GEDC01029207 GEDC01026752 GEDC01023081 GEDC01018861 GEDC01018035 GEDC01009433 JAS07170.1 JAS07234.1 JAS08091.1 JAS10546.1 JAS14217.1 JAS18437.1 JAS19263.1 JAS27865.1 AKCR02000021 PKK27127.1 KK611561 KFW12538.1 LSYS01005497 OPJ76896.1 GDIP01186095 JAJ37307.1 GDIP01149203 JAJ74199.1 GEBQ01020167 JAT19810.1 KL410243 KFQ94978.1 KQ434899 KZC10841.1 GDIP01145753 JAJ77649.1 GDIP01076240 LRGB01000337 JAM27475.1 KZS19664.1 GDIP01097106 JAM06609.1 KK537385 KFP29830.1 JARO02004996 KPP67527.1 KL449254 KFV04340.1 KL224841 KFW64799.1 KL502029 KFO14919.1 KL225950 KFM05260.1 KK735494 KFR14443.1 KL873593 KGL99557.1 HADZ01010046 HAEA01009165 SBQ37645.1 KK813906 KFQ37486.1 KK842910 KFP85057.1 KL416006 KFW81220.1 HAEF01002094 SBR39476.1 KK483599 KFQ61246.1 HAEJ01013197 SBS53654.1 AFYH01138846 AFYH01138847 KZ505751 PKU45604.1 HADY01009407 SBP47892.1 KK442277 KFQ70581.1 LMAW01000080 KQL60878.1 FR904263 CDQ56460.1 FR904630 CDQ68492.1 KL669406 KFW76315.1 GDIQ01103653 JAL48073.1 GDIP01022431 JAM81284.1 HAED01017793 SBR04238.1 JW871548 AFP04066.1 KK945584 KFQ54061.1 AYCK01010660 KL447609 KFO75190.1 KL216219 KFV68456.1 AHAT01039651 KL245483 KFV14512.1
SOQ47640.1 AGBW02012181 OWR45488.1 KQ458575 KPJ05687.1 KQ460772 KPJ12449.1 GAIX01013911 JAA78649.1 JTDY01009783 KOB62713.1 NEVH01002717 PNF41403.1 KK853253 KDR09309.1 GBYB01010908 GBYB01010909 JAG80675.1 JAG80676.1 KK107371 EZA51846.1 QOIP01000002 RLU25333.1 KQ976424 KYM88393.1 ADTU01012852 KQ981906 KYN33259.1 KQ977600 KYN01567.1 KZ288368 PBC26816.1 KQ982548 KYQ55198.1 GL768123 EFZ12079.1 KQ981153 KYN08890.1 KQ435851 KOX70729.1 KQ760629 OAD59737.1 BT081857 ACO51988.1 NNAY01000827 OXU26299.1 KK502178 KFP21325.1 KL357092 KFZ62944.1 CM012438 RVE75381.1 GEDC01030128 GEDC01030064 GEDC01029207 GEDC01026752 GEDC01023081 GEDC01018861 GEDC01018035 GEDC01009433 JAS07170.1 JAS07234.1 JAS08091.1 JAS10546.1 JAS14217.1 JAS18437.1 JAS19263.1 JAS27865.1 AKCR02000021 PKK27127.1 KK611561 KFW12538.1 LSYS01005497 OPJ76896.1 GDIP01186095 JAJ37307.1 GDIP01149203 JAJ74199.1 GEBQ01020167 JAT19810.1 KL410243 KFQ94978.1 KQ434899 KZC10841.1 GDIP01145753 JAJ77649.1 GDIP01076240 LRGB01000337 JAM27475.1 KZS19664.1 GDIP01097106 JAM06609.1 KK537385 KFP29830.1 JARO02004996 KPP67527.1 KL449254 KFV04340.1 KL224841 KFW64799.1 KL502029 KFO14919.1 KL225950 KFM05260.1 KK735494 KFR14443.1 KL873593 KGL99557.1 HADZ01010046 HAEA01009165 SBQ37645.1 KK813906 KFQ37486.1 KK842910 KFP85057.1 KL416006 KFW81220.1 HAEF01002094 SBR39476.1 KK483599 KFQ61246.1 HAEJ01013197 SBS53654.1 AFYH01138846 AFYH01138847 KZ505751 PKU45604.1 HADY01009407 SBP47892.1 KK442277 KFQ70581.1 LMAW01000080 KQL60878.1 FR904263 CDQ56460.1 FR904630 CDQ68492.1 KL669406 KFW76315.1 GDIQ01103653 JAL48073.1 GDIP01022431 JAM81284.1 HAED01017793 SBR04238.1 JW871548 AFP04066.1 KK945584 KFQ54061.1 AYCK01010660 KL447609 KFO75190.1 KL216219 KFV68456.1 AHAT01039651 KL245483 KFV14512.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000235965 UP000027135 UP000085678 UP000005203 UP000053097 UP000279307 UP000078540 UP000005205 UP000078541 UP000078542 UP000242457 UP000075809 UP000002358 UP000078492 UP000053105 UP000215335 UP000053119 UP000053872 UP000190648 UP000053283 UP000076502 UP000265180 UP000076858 UP000034805 UP000192224 UP000005226 UP000054081 UP000265200 UP000053286 UP000053605 UP000053858 UP000087266 UP000001038 UP000008672 UP000051836 UP000193380 UP000053258 UP000001646 UP000261560 UP000261480 UP000028760 UP000265140 UP000053760 UP000053875 UP000018468 UP000261460 UP000002852
UP000235965 UP000027135 UP000085678 UP000005203 UP000053097 UP000279307 UP000078540 UP000005205 UP000078541 UP000078542 UP000242457 UP000075809 UP000002358 UP000078492 UP000053105 UP000215335 UP000053119 UP000053872 UP000190648 UP000053283 UP000076502 UP000265180 UP000076858 UP000034805 UP000192224 UP000005226 UP000054081 UP000265200 UP000053286 UP000053605 UP000053858 UP000087266 UP000001038 UP000008672 UP000051836 UP000193380 UP000053258 UP000001646 UP000261560 UP000261480 UP000028760 UP000265140 UP000053760 UP000053875 UP000018468 UP000261460 UP000002852
PRIDE
Pfam
PF13092 CENP-L
Interpro
IPR025204
CENP-L
ProteinModelPortal
H9IXB1
A0A2A4K363
A0A2H1W3H5
A0A212EVG4
A0A194QJI5
A0A0N1I8H0
+ More
S4NNI8 A0A0L7KHB3 A0A2J7RKN5 A0A067QX65 A0A0C9Q978 A0A1S3K664 A0A2R2MSD2 A0A1S3H1I0 A0A1S3GZN3 A0A088AJ36 A0A026W870 A0A3L8DYN7 A0A195BQI1 A0A158NDS1 A0A195EZF6 A0A195CM69 A0A2A3E6G7 A0A151X4P6 K7ISD0 E9J519 A0A195D7M7 A0A0M8ZW33 A0A310SEQ0 C1C4S8 A0A232F773 A0A091JNI2 A0A094LG46 A0A3S2MGF1 A0A1B6CL68 A0A2I0MBS0 A0A093JF37 A0A1V4JXH2 A0A0P5BF54 A0A0P5EF24 A0A1B6L7Y6 A0A091VZG8 A0A154PG61 A0A0N8AHX3 A0A3P9L7G8 A0A0P5X9N8 A0A0P5WM11 A0A091JYJ4 A0A0P7WU96 A0A1W5A7N1 A0A093BKM9 A0A3B5KA17 A0A093R1B7 A0A087VQM7 A0A3P9JHC3 A0A087QVK6 A0A091WI06 A0A0A0B049 A0A1A8DTH9 A0A091RD97 A0A1S3R182 A0A091N7S0 A0A093Q463 H2LJ76 A0A1A8L523 A0A091TRF0 A0A1A8UZ18 H3AUJ6 A0A2I0UHS7 A0A1A7ZYB5 A0A091T855 A0A0Q3U511 A0A060VVV0 A0A060WN69 A0A093PPY7 H2TXD4 A0A0P5RAL4 A0A0N8DLC7 G1KR38 A0A1A8J6B7 A0A3B3BHQ5 A0A3B3Y555 V9KYT9 A0A091SAU5 A0A087X579 A0A3P8Y0F4 A0A091FY01 A0A093IP24 W5MS61 A0A093CQC8 A0A3B4H300 A0A3B5QRX3
S4NNI8 A0A0L7KHB3 A0A2J7RKN5 A0A067QX65 A0A0C9Q978 A0A1S3K664 A0A2R2MSD2 A0A1S3H1I0 A0A1S3GZN3 A0A088AJ36 A0A026W870 A0A3L8DYN7 A0A195BQI1 A0A158NDS1 A0A195EZF6 A0A195CM69 A0A2A3E6G7 A0A151X4P6 K7ISD0 E9J519 A0A195D7M7 A0A0M8ZW33 A0A310SEQ0 C1C4S8 A0A232F773 A0A091JNI2 A0A094LG46 A0A3S2MGF1 A0A1B6CL68 A0A2I0MBS0 A0A093JF37 A0A1V4JXH2 A0A0P5BF54 A0A0P5EF24 A0A1B6L7Y6 A0A091VZG8 A0A154PG61 A0A0N8AHX3 A0A3P9L7G8 A0A0P5X9N8 A0A0P5WM11 A0A091JYJ4 A0A0P7WU96 A0A1W5A7N1 A0A093BKM9 A0A3B5KA17 A0A093R1B7 A0A087VQM7 A0A3P9JHC3 A0A087QVK6 A0A091WI06 A0A0A0B049 A0A1A8DTH9 A0A091RD97 A0A1S3R182 A0A091N7S0 A0A093Q463 H2LJ76 A0A1A8L523 A0A091TRF0 A0A1A8UZ18 H3AUJ6 A0A2I0UHS7 A0A1A7ZYB5 A0A091T855 A0A0Q3U511 A0A060VVV0 A0A060WN69 A0A093PPY7 H2TXD4 A0A0P5RAL4 A0A0N8DLC7 G1KR38 A0A1A8J6B7 A0A3B3BHQ5 A0A3B3Y555 V9KYT9 A0A091SAU5 A0A087X579 A0A3P8Y0F4 A0A091FY01 A0A093IP24 W5MS61 A0A093CQC8 A0A3B4H300 A0A3B5QRX3
Ontologies
GO
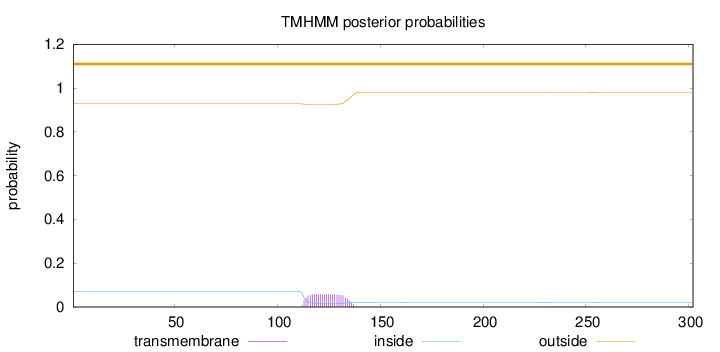
Topology
Length:
302
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.28911
Exp number, first 60 AAs:
0.00051
Total prob of N-in:
0.07193
outside
1 - 302
Population Genetic Test Statistics
Pi
299.664836
Theta
200.758707
Tajima's D
1.358226
CLR
0.003973
CSRT
0.754962251887406
Interpretation
Uncertain
