Gene
KWMTBOMO11444
Pre Gene Modal
BGIBMGA001930
Annotation
PREDICTED:_growth_arrest-specific_protein_1-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.727
Sequence
CDS
ATGTGGGTGGTGGCCGCGTTGCTGGCGGCATCGGCGGCGGCGGTGGCGTCCATGCCGTGCACGGAGGCCCGCAACCGCTGCATGTACCGGACCGGATGCGGCGCCGCGCTCTCCAACTACATGATGATGTGCGAACGAGTGCTCACAACCGACTTGCCCCCCACGGCATGCCCAAACGAATGCAGCAACGCTCTTATCGCGCTCACTTCCACCGAAGAGGGAAAGGAGCTCATGAGTTGTGAATGCGAAGACGACTATTGCATAGAGGCAAAGGACAGGATTGACGTGTGTCGGTCACAAGTAATGAAGGGCGCTTCGGATGTCATACCATCCTGCAGCCTCTCCCAACTAATATGCCTGGCCGACGCCCAGTGTTCGACTGCTTTACAATACTACCATCATCTATGCCGTTCAATGTTCCGAGGAAGAAAATGCTCAAAAAAATGCATTAACTCCATTGAAATTCTAAGGAAACAGGAAAAGGCCGCAGCGTTGACAGTTTGCCAATGCGACGGCACCGAAGACTACGATTGCCCTACAATGCAAGAGAATCTGGCTAGGTTGTGCTTCCACAAACATAAGAATCACACGCGAAGTCATGACCGACACGGAGAGAAACATAAGAAAGTCAATGAAGTAGTTTATGCGAGTGCTTCCAGTCTCAGTGATATATCGCTGTTGTTATTCTTGACTTGTATTCTGAGTGCTTTCTCATGA
Protein
MWVVAALLAASAAAVASMPCTEARNRCMYRTGCGAALSNYMMMCERVLTTDLPPTACPNECSNALIALTSTEEGKELMSCECEDDYCIEAKDRIDVCRSQVMKGASDVIPSCSLSQLICLADAQCSTALQYYHHLCRSMFRGRKCSKKCINSIEILRKQEKAAALTVCQCDGTEDYDCPTMQENLARLCFHKHKNHTRSHDRHGEKHKKVNEVVYASASSLSDISLLLFLTCILSAFS
Summary
Similarity
Belongs to the peptidase S8 family.
Uniprot
H9IXE7
A0A2A4K2D9
A0A2H1WS76
A0A0N1PEF9
A0A194RAV2
A0A3S2NDI8
+ More
D6WS04 A0A1Y1N4K2 V5H417 A0A2J7R7X0 A0A2P8YNJ3 A0A1B6MJI7 T1HBC4 A0A1W4WMS4 E9FRQ2 A0A0P5GQX2 A0A0P6BJ36 A0A0A9WVU9 A0A0P6HWN4 A0A0K8SBP6 A0A154PHD5 A0A0L7RH79 U4UFE9 N6T3A5 A0A0M9A4V8 A0A2A3ERN4 A0A088ASB4 A0A0T6BBZ5 A0A2P6KL70 A0A0C9QKG8 A0A087TDM8 A0A0K2TE10 A0A2L2YIA9 A0A2H8TJJ6 T1IMB0 A0A2S2PLA4 J9JJI6 A0A2S2PWM9 A0A2C9JJV4 A0A0K2USK2 C1BUK8 A0A0B6ZG99 A0A0L8FF55 A0A210QVT1 V4B1P8 A0A2T7NFL7 R7US30 R7V4L7 A0A1W0X5C4 K1QZZ3 W5LNL5 A0A0S7HT31 A0A3P9NTW2 A0A2U9BDS8 A0A3B3YS52 A0A3B3VCD3 A0A087YS01 A0A3Q2X8H0 A0A3P9LCY6 V9L834 A0A3B3TFR2 A0A3Q1HEQ4 A0A3Q2NW07 M4B109 A0A3B5LP82 A0A147A6L3 A0A146WR48 H2ZTL1 Q08CY8 F6S7D7 A0A3P9JL82 H2MH78 A0A2D0QU48 A0A3B4XZA3 A0A3B4TWI0 A0A146R990 A0A3Q3RRC3 H3A7W6 A0A3P8SGQ1 A0A3N0Y7M1 A0A3Q4BQ87 A0A3Q1F7F9 A0A3B3ZV03 A0A3Q2CWD7 A0A3Q3JLC3 A0A1L8HYD8
D6WS04 A0A1Y1N4K2 V5H417 A0A2J7R7X0 A0A2P8YNJ3 A0A1B6MJI7 T1HBC4 A0A1W4WMS4 E9FRQ2 A0A0P5GQX2 A0A0P6BJ36 A0A0A9WVU9 A0A0P6HWN4 A0A0K8SBP6 A0A154PHD5 A0A0L7RH79 U4UFE9 N6T3A5 A0A0M9A4V8 A0A2A3ERN4 A0A088ASB4 A0A0T6BBZ5 A0A2P6KL70 A0A0C9QKG8 A0A087TDM8 A0A0K2TE10 A0A2L2YIA9 A0A2H8TJJ6 T1IMB0 A0A2S2PLA4 J9JJI6 A0A2S2PWM9 A0A2C9JJV4 A0A0K2USK2 C1BUK8 A0A0B6ZG99 A0A0L8FF55 A0A210QVT1 V4B1P8 A0A2T7NFL7 R7US30 R7V4L7 A0A1W0X5C4 K1QZZ3 W5LNL5 A0A0S7HT31 A0A3P9NTW2 A0A2U9BDS8 A0A3B3YS52 A0A3B3VCD3 A0A087YS01 A0A3Q2X8H0 A0A3P9LCY6 V9L834 A0A3B3TFR2 A0A3Q1HEQ4 A0A3Q2NW07 M4B109 A0A3B5LP82 A0A147A6L3 A0A146WR48 H2ZTL1 Q08CY8 F6S7D7 A0A3P9JL82 H2MH78 A0A2D0QU48 A0A3B4XZA3 A0A3B4TWI0 A0A146R990 A0A3Q3RRC3 H3A7W6 A0A3P8SGQ1 A0A3N0Y7M1 A0A3Q4BQ87 A0A3Q1F7F9 A0A3B3ZV03 A0A3Q2CWD7 A0A3Q3JLC3 A0A1L8HYD8
Pubmed
EMBL
BABH01018032
NWSH01000239
PCG78098.1
ODYU01010602
SOQ55822.1
KQ459169
+ More
KPJ03417.1 KQ460436 KPJ14732.1 RSAL01000086 RVE48318.1 KQ971351 EFA06406.1 GEZM01013038 JAV92802.1 GALX01000918 JAB67548.1 NEVH01006723 PNF36924.1 PYGN01000467 PSN45828.1 GEBQ01003922 JAT36055.1 ACPB03009220 GL732523 EFX90446.1 GDIQ01260286 LRGB01000592 JAJ91438.1 KZS17738.1 GDIP01028352 JAM75363.1 GBHO01034614 JAG08990.1 GDIQ01014423 JAN80314.1 GBRD01015636 GDHC01018000 JAG50190.1 JAQ00629.1 KQ434905 KZC11265.1 KQ414592 KOC70193.1 KB632085 ERL88640.1 APGK01045482 KB741037 ENN74604.1 KQ435756 KOX75983.1 KZ288191 PBC34425.1 LJIG01002068 KRT84872.1 MWRG01009443 PRD27076.1 GBYB01004059 JAG73826.1 KK114744 KFM63217.1 HACA01006330 CDW23691.1 IAAA01023018 IAAA01023019 LAA07849.1 GFXV01001310 GFXV01002489 MBW13115.1 MBW14294.1 JH431009 GGMR01017584 MBY30203.1 ABLF02021517 GGMS01000561 MBY69764.1 HACA01023893 CDW41254.1 BT078287 ACO12711.1 HACG01019895 CEK66760.1 KQ433944 KOF62290.1 NEDP02001616 OWF52860.1 KB200521 ESP01246.1 PZQS01000013 PVD19969.1 AMQN01007332 KB300467 ELU06717.1 AMQN01005144 KB295236 ELU13412.1 MTYJ01000016 OQV22683.1 JH816980 EKC42622.1 GBYX01437236 JAO44111.1 CP026247 AWP01939.1 AYCK01007181 JW875051 AFP07568.1 GCES01012172 JAR74151.1 GCES01053992 JAR32331.1 AFYH01270921 BC124029 AAI24030.1 AAMC01039569 GCES01121401 JAQ64921.1 AFYH01054001 RJVU01050255 ROL42159.1 CM004466 OCU01160.1
KPJ03417.1 KQ460436 KPJ14732.1 RSAL01000086 RVE48318.1 KQ971351 EFA06406.1 GEZM01013038 JAV92802.1 GALX01000918 JAB67548.1 NEVH01006723 PNF36924.1 PYGN01000467 PSN45828.1 GEBQ01003922 JAT36055.1 ACPB03009220 GL732523 EFX90446.1 GDIQ01260286 LRGB01000592 JAJ91438.1 KZS17738.1 GDIP01028352 JAM75363.1 GBHO01034614 JAG08990.1 GDIQ01014423 JAN80314.1 GBRD01015636 GDHC01018000 JAG50190.1 JAQ00629.1 KQ434905 KZC11265.1 KQ414592 KOC70193.1 KB632085 ERL88640.1 APGK01045482 KB741037 ENN74604.1 KQ435756 KOX75983.1 KZ288191 PBC34425.1 LJIG01002068 KRT84872.1 MWRG01009443 PRD27076.1 GBYB01004059 JAG73826.1 KK114744 KFM63217.1 HACA01006330 CDW23691.1 IAAA01023018 IAAA01023019 LAA07849.1 GFXV01001310 GFXV01002489 MBW13115.1 MBW14294.1 JH431009 GGMR01017584 MBY30203.1 ABLF02021517 GGMS01000561 MBY69764.1 HACA01023893 CDW41254.1 BT078287 ACO12711.1 HACG01019895 CEK66760.1 KQ433944 KOF62290.1 NEDP02001616 OWF52860.1 KB200521 ESP01246.1 PZQS01000013 PVD19969.1 AMQN01007332 KB300467 ELU06717.1 AMQN01005144 KB295236 ELU13412.1 MTYJ01000016 OQV22683.1 JH816980 EKC42622.1 GBYX01437236 JAO44111.1 CP026247 AWP01939.1 AYCK01007181 JW875051 AFP07568.1 GCES01012172 JAR74151.1 GCES01053992 JAR32331.1 AFYH01270921 BC124029 AAI24030.1 AAMC01039569 GCES01121401 JAQ64921.1 AFYH01054001 RJVU01050255 ROL42159.1 CM004466 OCU01160.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007266
+ More
UP000235965 UP000245037 UP000015103 UP000192223 UP000000305 UP000076858 UP000076502 UP000053825 UP000030742 UP000019118 UP000053105 UP000242457 UP000005203 UP000054359 UP000007819 UP000076420 UP000053454 UP000242188 UP000030746 UP000245119 UP000014760 UP000005408 UP000018467 UP000242638 UP000246464 UP000261480 UP000261500 UP000028760 UP000264820 UP000265180 UP000261540 UP000265040 UP000265000 UP000002852 UP000261380 UP000008672 UP000008143 UP000265200 UP000001038 UP000221080 UP000261360 UP000261420 UP000261640 UP000265080 UP000261620 UP000257200 UP000261520 UP000265020 UP000261600 UP000186698
UP000235965 UP000245037 UP000015103 UP000192223 UP000000305 UP000076858 UP000076502 UP000053825 UP000030742 UP000019118 UP000053105 UP000242457 UP000005203 UP000054359 UP000007819 UP000076420 UP000053454 UP000242188 UP000030746 UP000245119 UP000014760 UP000005408 UP000018467 UP000242638 UP000246464 UP000261480 UP000261500 UP000028760 UP000264820 UP000265180 UP000261540 UP000265040 UP000265000 UP000002852 UP000261380 UP000008672 UP000008143 UP000265200 UP000001038 UP000221080 UP000261360 UP000261420 UP000261640 UP000265080 UP000261620 UP000257200 UP000261520 UP000265020 UP000261600 UP000186698
PRIDE
Pfam
Interpro
IPR039596
GAS1
+ More
IPR016017 GDNF/GAS1
IPR015500 Peptidase_S8_subtilisin-rel
IPR038466 S8_pro-domain_sf
IPR013579 FAST_2
IPR000209 Peptidase_S8/S53_dom
IPR008979 Galactose-bd-like_sf
IPR036852 Peptidase_S8/S53_dom_sf
IPR006212 Furin_repeat
IPR014848 Rgp1
IPR023827 Peptidase_S8_Asp-AS
IPR002884 P_dom
IPR007632 Anoctamin
IPR009030 Growth_fac_rcpt_cys_sf
IPR022398 Peptidase_S8_His-AS
IPR032815 S8_pro-domain
IPR034182 Kexin/furin
IPR013584 RAP
IPR023828 Peptidase_S8_Ser-AS
IPR037193 GDNF_alpha
IPR016017 GDNF/GAS1
IPR015500 Peptidase_S8_subtilisin-rel
IPR038466 S8_pro-domain_sf
IPR013579 FAST_2
IPR000209 Peptidase_S8/S53_dom
IPR008979 Galactose-bd-like_sf
IPR036852 Peptidase_S8/S53_dom_sf
IPR006212 Furin_repeat
IPR014848 Rgp1
IPR023827 Peptidase_S8_Asp-AS
IPR002884 P_dom
IPR007632 Anoctamin
IPR009030 Growth_fac_rcpt_cys_sf
IPR022398 Peptidase_S8_His-AS
IPR032815 S8_pro-domain
IPR034182 Kexin/furin
IPR013584 RAP
IPR023828 Peptidase_S8_Ser-AS
IPR037193 GDNF_alpha
Gene 3D
ProteinModelPortal
H9IXE7
A0A2A4K2D9
A0A2H1WS76
A0A0N1PEF9
A0A194RAV2
A0A3S2NDI8
+ More
D6WS04 A0A1Y1N4K2 V5H417 A0A2J7R7X0 A0A2P8YNJ3 A0A1B6MJI7 T1HBC4 A0A1W4WMS4 E9FRQ2 A0A0P5GQX2 A0A0P6BJ36 A0A0A9WVU9 A0A0P6HWN4 A0A0K8SBP6 A0A154PHD5 A0A0L7RH79 U4UFE9 N6T3A5 A0A0M9A4V8 A0A2A3ERN4 A0A088ASB4 A0A0T6BBZ5 A0A2P6KL70 A0A0C9QKG8 A0A087TDM8 A0A0K2TE10 A0A2L2YIA9 A0A2H8TJJ6 T1IMB0 A0A2S2PLA4 J9JJI6 A0A2S2PWM9 A0A2C9JJV4 A0A0K2USK2 C1BUK8 A0A0B6ZG99 A0A0L8FF55 A0A210QVT1 V4B1P8 A0A2T7NFL7 R7US30 R7V4L7 A0A1W0X5C4 K1QZZ3 W5LNL5 A0A0S7HT31 A0A3P9NTW2 A0A2U9BDS8 A0A3B3YS52 A0A3B3VCD3 A0A087YS01 A0A3Q2X8H0 A0A3P9LCY6 V9L834 A0A3B3TFR2 A0A3Q1HEQ4 A0A3Q2NW07 M4B109 A0A3B5LP82 A0A147A6L3 A0A146WR48 H2ZTL1 Q08CY8 F6S7D7 A0A3P9JL82 H2MH78 A0A2D0QU48 A0A3B4XZA3 A0A3B4TWI0 A0A146R990 A0A3Q3RRC3 H3A7W6 A0A3P8SGQ1 A0A3N0Y7M1 A0A3Q4BQ87 A0A3Q1F7F9 A0A3B3ZV03 A0A3Q2CWD7 A0A3Q3JLC3 A0A1L8HYD8
D6WS04 A0A1Y1N4K2 V5H417 A0A2J7R7X0 A0A2P8YNJ3 A0A1B6MJI7 T1HBC4 A0A1W4WMS4 E9FRQ2 A0A0P5GQX2 A0A0P6BJ36 A0A0A9WVU9 A0A0P6HWN4 A0A0K8SBP6 A0A154PHD5 A0A0L7RH79 U4UFE9 N6T3A5 A0A0M9A4V8 A0A2A3ERN4 A0A088ASB4 A0A0T6BBZ5 A0A2P6KL70 A0A0C9QKG8 A0A087TDM8 A0A0K2TE10 A0A2L2YIA9 A0A2H8TJJ6 T1IMB0 A0A2S2PLA4 J9JJI6 A0A2S2PWM9 A0A2C9JJV4 A0A0K2USK2 C1BUK8 A0A0B6ZG99 A0A0L8FF55 A0A210QVT1 V4B1P8 A0A2T7NFL7 R7US30 R7V4L7 A0A1W0X5C4 K1QZZ3 W5LNL5 A0A0S7HT31 A0A3P9NTW2 A0A2U9BDS8 A0A3B3YS52 A0A3B3VCD3 A0A087YS01 A0A3Q2X8H0 A0A3P9LCY6 V9L834 A0A3B3TFR2 A0A3Q1HEQ4 A0A3Q2NW07 M4B109 A0A3B5LP82 A0A147A6L3 A0A146WR48 H2ZTL1 Q08CY8 F6S7D7 A0A3P9JL82 H2MH78 A0A2D0QU48 A0A3B4XZA3 A0A3B4TWI0 A0A146R990 A0A3Q3RRC3 H3A7W6 A0A3P8SGQ1 A0A3N0Y7M1 A0A3Q4BQ87 A0A3Q1F7F9 A0A3B3ZV03 A0A3Q2CWD7 A0A3Q3JLC3 A0A1L8HYD8
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.990148
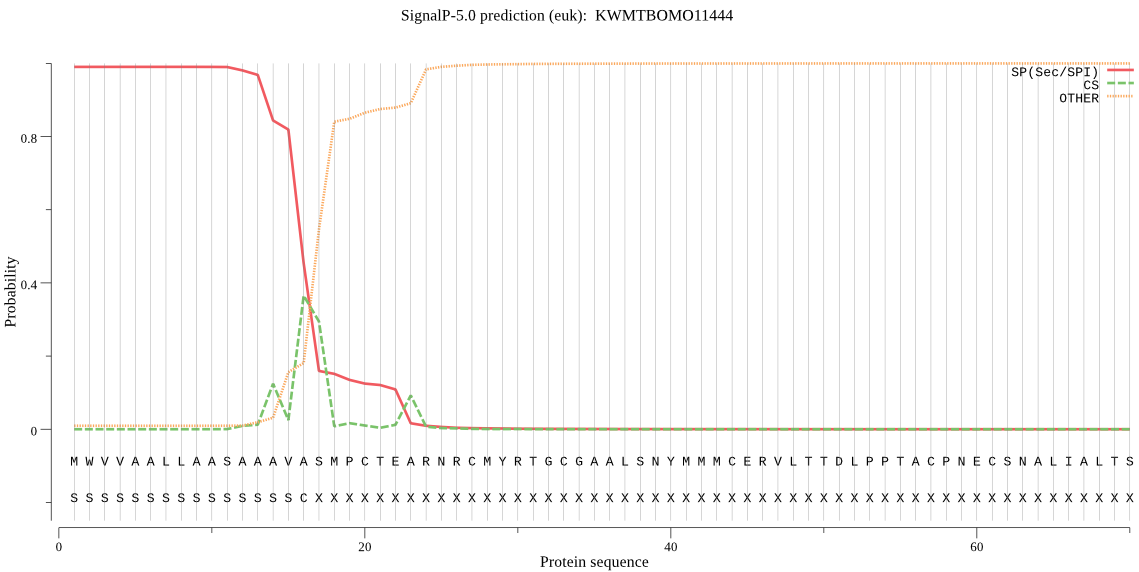
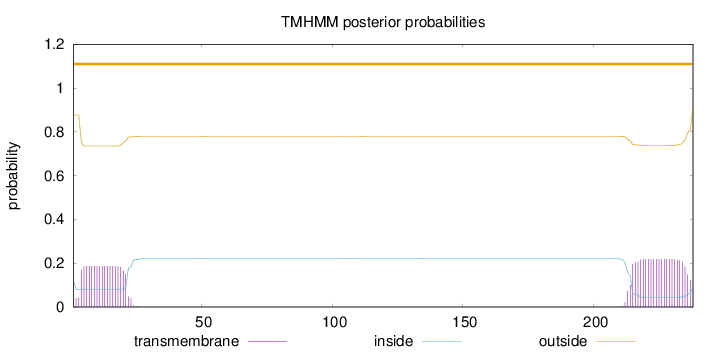
Length:
238
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.43542000000001
Exp number, first 60 AAs:
3.47239
Total prob of N-in:
0.12290
outside
1 - 238
Population Genetic Test Statistics
Pi
160.71049
Theta
150.551557
Tajima's D
0.043166
CLR
106.908597
CSRT
0.378181090945453
Interpretation
Uncertain
