Gene
KWMTBOMO11432
Pre Gene Modal
BGIBMGA001938
Annotation
PREDICTED:_probable_protein_arginine_N-methyltransferase_6_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 3.079
Sequence
CDS
ATGGAAACTGAAGAAGTGCAGAACAATTACTTTAACAGCTATGAAGATTTAGAGGTGCATACATTGATGTTAGAAGACGAACCAAGAAACCTAGCTTATAAGAAAGCGATATTAGGGAATAAATGTTATTTCAAGGATAAAATAGTAATGGATGTTGGATGTGGCACAGGAATTTTATCTGTTTTCTGCGCCCAAGCAGATGCTAAAAAAGTATATGCCGTGGAAGCAAGTAATTTAGCGAATTTAGCTAGAGAAATTGTGAAAGAGAATGGTTTCGAAAATGTTATTGAAATAATTCACAGTAAAGTTGAAGATGTAATATTACCAAATAACATAAAAGTAGATGTGATAGTATCTGAATGGATGGGTTTTTATCTATTGCATGAAGGTATGTTAGATTCTGTGCTAATTGCTCGAGATAAGTTTCTTAAGGAAGGGGGAGAAATGTTTCCTGAAACGGCTGTTATTTATGTTGCACCATGTAGCATACCTTCACTATACAACAAATGGAACAATGTGCATGGAGTATCAATGTCTGCTTTTGCTAAACATCTTCGCAAAAACAAATCCACTAAACCAGAAATTTTGAATATAGAGCAAAAGGATTTGCTTGGTGATGAAGTAGCACTTTGCTGGATTAATTTGAGGGAAGACAGTGCATCTGATTTGAATGAATTCAGCATCCAGCATGTTGTTGGAGCTAAAAAATCTGGTTCTTATGAAGGCTTGTGTCTATGGTTCGAATGCAATTTTCCCAGTATGTCTGAAACGAAAGACGAGAATCGTATAATTTTGGACACCAGTCCAAATAGTTTGGCAACACACTGGAAACAAACAGCAATAGTATTACCACAGGAAGTGGAAGTGGAAGAAGGAGAACCAATAGCTTTCCAACTCAGCATGAAAAGGGATACAGAACATAACCGGAGATACAATTTACAGATGACATTACTGGATCCAGAAGCTGTCGATCACCCACAACCATGCTCATGTCACATGACTAAATGCATACTGATTAAAGCATTCATGGAGTCCCATTAA
Protein
METEEVQNNYFNSYEDLEVHTLMLEDEPRNLAYKKAILGNKCYFKDKIVMDVGCGTGILSVFCAQADAKKVYAVEASNLANLAREIVKENGFENVIEIIHSKVEDVILPNNIKVDVIVSEWMGFYLLHEGMLDSVLIARDKFLKEGGEMFPETAVIYVAPCSIPSLYNKWNNVHGVSMSAFAKHLRKNKSTKPEILNIEQKDLLGDEVALCWINLREDSASDLNEFSIQHVVGAKKSGSYEGLCLWFECNFPSMSETKDENRIILDTSPNSLATHWKQTAIVLPQEVEVEEGEPIAFQLSMKRDTEHNRRYNLQMTLLDPEAVDHPQPCSCHMTKCILIKAFMESH
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. Protein arginine N-methyltransferase family.
Uniprot
A0A2H1V472
S4PTE8
H9IXF5
A0A2Z5UIW9
A0A2A4K697
A0A212FGZ3
+ More
A0A3S2M8A6 A0A194QKG5 A0A194RAW8 Q0IG24 A0A084VYH0 A0A1B3PD61 A0A182QPS2 A0A182H4C5 N6TZA5 J3JT85 Q7PWU7 A0A1S4GZE9 A0A182XDI6 A0A1B3PDI8 A0A182KNH0 A0A182VJW1 A0A182NUP0 A0A2M4ASN9 B4P156 A0A182MAC8 A0A0J9R038 A0A1A9YH44 A0A2M4AS86 Q8SX32 B4HWZ7 W5JVJ4 A0A182UHS3 A0A1B0C0R2 A0A1B0FCW3 A0A0T6ATR6 A0A1A9ZEY4 A0A1B3PDG5 A0A182I732 A0A1I8N6K0 B3N4Z5 A0A139WG38 B3MK00 A0A182VU60 A0A182Y972 T1DTM4 A0A2M4BSV7 A0A182P9Q1 A0A1A9WLY0 A0A182J705 A0A2M4BSV1 A0A182R4A0 B4KHQ5 A0A1W4VSY3 A0A182K391 B4LVB2 A0A3B0K6Q8 A0A2M3ZBC0 A0A2M3ZB12 A0A1J1IXV1 A0A182F1Z2 B4JQ09 A0A0M4EQJ1 A0A182SXK9 A0A1B6IYR1 B0WU19 A0A2J7R7Y1 A0A2J7R7W6 A0A2M4ASL5 F4WX56 A0A1B6DBT6 A0A1A9VI55 A0A1B0DD49 A0A195FAA6 A0A2P8XI50 A0A1B6KTC2 A0A1Y1MT54 A0A232FIH2 A0A158NCJ5 E2ALX0 A0A195EMT1 A0A1L8DM44 A0A1B0GL87 E9IM91 A0A195BWH1 A0A336LS43 A0A0P5CQZ0 A0A154P2Z9 A0A0N8CF82 A0A195CS44 B4QA90 A0A0P5FS48 K7IQH8 J9JST9 B4MUA4 A0A2S2QSN3 A0A0N8B5X4 A0A0P5IW72 A0A2S2PIV1 A0A0P5CXL8
A0A3S2M8A6 A0A194QKG5 A0A194RAW8 Q0IG24 A0A084VYH0 A0A1B3PD61 A0A182QPS2 A0A182H4C5 N6TZA5 J3JT85 Q7PWU7 A0A1S4GZE9 A0A182XDI6 A0A1B3PDI8 A0A182KNH0 A0A182VJW1 A0A182NUP0 A0A2M4ASN9 B4P156 A0A182MAC8 A0A0J9R038 A0A1A9YH44 A0A2M4AS86 Q8SX32 B4HWZ7 W5JVJ4 A0A182UHS3 A0A1B0C0R2 A0A1B0FCW3 A0A0T6ATR6 A0A1A9ZEY4 A0A1B3PDG5 A0A182I732 A0A1I8N6K0 B3N4Z5 A0A139WG38 B3MK00 A0A182VU60 A0A182Y972 T1DTM4 A0A2M4BSV7 A0A182P9Q1 A0A1A9WLY0 A0A182J705 A0A2M4BSV1 A0A182R4A0 B4KHQ5 A0A1W4VSY3 A0A182K391 B4LVB2 A0A3B0K6Q8 A0A2M3ZBC0 A0A2M3ZB12 A0A1J1IXV1 A0A182F1Z2 B4JQ09 A0A0M4EQJ1 A0A182SXK9 A0A1B6IYR1 B0WU19 A0A2J7R7Y1 A0A2J7R7W6 A0A2M4ASL5 F4WX56 A0A1B6DBT6 A0A1A9VI55 A0A1B0DD49 A0A195FAA6 A0A2P8XI50 A0A1B6KTC2 A0A1Y1MT54 A0A232FIH2 A0A158NCJ5 E2ALX0 A0A195EMT1 A0A1L8DM44 A0A1B0GL87 E9IM91 A0A195BWH1 A0A336LS43 A0A0P5CQZ0 A0A154P2Z9 A0A0N8CF82 A0A195CS44 B4QA90 A0A0P5FS48 K7IQH8 J9JST9 B4MUA4 A0A2S2QSN3 A0A0N8B5X4 A0A0P5IW72 A0A2S2PIV1 A0A0P5CXL8
Pubmed
23622113
19121390
22118469
26354079
17510324
24438588
+ More
26483478 23537049 22516182 12364791 20966253 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25315136 18362917 19820115 25244985 21719571 29403074 28004739 28648823 21347285 20798317 21282665 20075255
26483478 23537049 22516182 12364791 20966253 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25315136 18362917 19820115 25244985 21719571 29403074 28004739 28648823 21347285 20798317 21282665 20075255
EMBL
ODYU01000592
SOQ35645.1
GAIX01012163
JAA80397.1
BABH01018071
BABH01018072
+ More
AP017509 BBB06822.1 NWSH01000136 PCG79190.1 AGBW02008579 OWR53005.1 RSAL01000010 RVE53750.1 KQ459053 KPJ03951.1 KQ460436 KPJ14747.1 CH477274 EAT45189.1 ATLV01018366 KE525231 KFB43014.1 KU659866 AOG17665.1 AXCN02000736 JXUM01001217 APGK01045489 APGK01045490 KB741037 KB632085 ENN74605.1 ERL88638.1 BT126441 AEE61405.1 AAAB01008984 EAA14811.4 KU659978 AOG17776.1 GGFK01010459 MBW43780.1 CM000157 EDW88031.1 AXCM01000043 CM002910 KMY89677.1 GGFK01010322 MBW43643.1 AE014134 AY094881 AAF53052.2 AAM11234.1 CH480818 EDW52542.1 ADMH02000358 ETN66844.1 JXJN01023734 CCAG010017495 LJIG01022820 KRT78547.1 KU659976 AOG17774.1 APCN01003350 CH954177 EDV58940.1 KQ971351 KYB26849.1 CH902620 EDV32455.1 GAMD01000864 JAB00727.1 GGFJ01007024 MBW56165.1 GGFJ01007025 MBW56166.1 CH933807 EDW13342.1 CH940649 EDW63291.1 OUUW01000006 SPP81699.1 GGFM01005024 MBW25775.1 GGFM01004968 MBW25719.1 CVRI01000063 CRL04396.1 CH916372 EDV98989.1 CP012523 ALC39287.1 GECU01015667 JAS92039.1 DS232099 EDS34735.1 NEVH01006723 PNF36926.1 PNF36927.1 GGFK01010439 MBW43760.1 GL888417 EGI61299.1 GEDC01014149 JAS23149.1 AJVK01031635 KQ981727 KYN36979.1 PYGN01002049 PSN31688.1 GEBQ01025278 JAT14699.1 GEZM01028004 GEZM01028003 JAV86527.1 NNAY01000172 OXU30320.1 ADTU01011943 GL440682 EFN65562.1 KQ978691 KYN29194.1 GFDF01006603 JAV07481.1 AJWK01034545 GL764129 EFZ18527.1 KQ976396 KYM92979.1 UFQS01000134 UFQT01000134 SSX00255.1 SSX20635.1 GDIP01182743 JAJ40659.1 KQ434809 KZC06251.1 GDIP01137591 LRGB01001937 JAL66123.1 KZS09900.1 KQ977408 KYN02934.1 CM000361 EDX04645.1 GDIQ01258617 JAJ93107.1 ABLF02039149 CH963857 EDW76030.2 GGMS01011576 MBY80779.1 GDIQ01209667 JAK42058.1 GDIQ01209666 JAK42059.1 GGMR01016708 MBY29327.1 GDIP01163997 JAJ59405.1
AP017509 BBB06822.1 NWSH01000136 PCG79190.1 AGBW02008579 OWR53005.1 RSAL01000010 RVE53750.1 KQ459053 KPJ03951.1 KQ460436 KPJ14747.1 CH477274 EAT45189.1 ATLV01018366 KE525231 KFB43014.1 KU659866 AOG17665.1 AXCN02000736 JXUM01001217 APGK01045489 APGK01045490 KB741037 KB632085 ENN74605.1 ERL88638.1 BT126441 AEE61405.1 AAAB01008984 EAA14811.4 KU659978 AOG17776.1 GGFK01010459 MBW43780.1 CM000157 EDW88031.1 AXCM01000043 CM002910 KMY89677.1 GGFK01010322 MBW43643.1 AE014134 AY094881 AAF53052.2 AAM11234.1 CH480818 EDW52542.1 ADMH02000358 ETN66844.1 JXJN01023734 CCAG010017495 LJIG01022820 KRT78547.1 KU659976 AOG17774.1 APCN01003350 CH954177 EDV58940.1 KQ971351 KYB26849.1 CH902620 EDV32455.1 GAMD01000864 JAB00727.1 GGFJ01007024 MBW56165.1 GGFJ01007025 MBW56166.1 CH933807 EDW13342.1 CH940649 EDW63291.1 OUUW01000006 SPP81699.1 GGFM01005024 MBW25775.1 GGFM01004968 MBW25719.1 CVRI01000063 CRL04396.1 CH916372 EDV98989.1 CP012523 ALC39287.1 GECU01015667 JAS92039.1 DS232099 EDS34735.1 NEVH01006723 PNF36926.1 PNF36927.1 GGFK01010439 MBW43760.1 GL888417 EGI61299.1 GEDC01014149 JAS23149.1 AJVK01031635 KQ981727 KYN36979.1 PYGN01002049 PSN31688.1 GEBQ01025278 JAT14699.1 GEZM01028004 GEZM01028003 JAV86527.1 NNAY01000172 OXU30320.1 ADTU01011943 GL440682 EFN65562.1 KQ978691 KYN29194.1 GFDF01006603 JAV07481.1 AJWK01034545 GL764129 EFZ18527.1 KQ976396 KYM92979.1 UFQS01000134 UFQT01000134 SSX00255.1 SSX20635.1 GDIP01182743 JAJ40659.1 KQ434809 KZC06251.1 GDIP01137591 LRGB01001937 JAL66123.1 KZS09900.1 KQ977408 KYN02934.1 CM000361 EDX04645.1 GDIQ01258617 JAJ93107.1 ABLF02039149 CH963857 EDW76030.2 GGMS01011576 MBY80779.1 GDIQ01209667 JAK42058.1 GDIQ01209666 JAK42059.1 GGMR01016708 MBY29327.1 GDIP01163997 JAJ59405.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000008820 UP000030765 UP000075886 UP000069940 UP000019118 UP000030742 UP000007062 UP000076407 UP000075882 UP000075903 UP000075884 UP000002282 UP000075883 UP000092443 UP000000803 UP000001292 UP000000673 UP000075902 UP000092460 UP000092444 UP000092445 UP000075840 UP000095301 UP000008711 UP000007266 UP000007801 UP000075920 UP000076408 UP000075885 UP000091820 UP000075880 UP000075900 UP000009192 UP000192221 UP000075881 UP000008792 UP000268350 UP000183832 UP000069272 UP000001070 UP000092553 UP000075901 UP000002320 UP000235965 UP000007755 UP000078200 UP000092462 UP000078541 UP000245037 UP000215335 UP000005205 UP000000311 UP000078492 UP000092461 UP000078540 UP000076502 UP000076858 UP000078542 UP000000304 UP000002358 UP000007819 UP000007798
UP000008820 UP000030765 UP000075886 UP000069940 UP000019118 UP000030742 UP000007062 UP000076407 UP000075882 UP000075903 UP000075884 UP000002282 UP000075883 UP000092443 UP000000803 UP000001292 UP000000673 UP000075902 UP000092460 UP000092444 UP000092445 UP000075840 UP000095301 UP000008711 UP000007266 UP000007801 UP000075920 UP000076408 UP000075885 UP000091820 UP000075880 UP000075900 UP000009192 UP000192221 UP000075881 UP000008792 UP000268350 UP000183832 UP000069272 UP000001070 UP000092553 UP000075901 UP000002320 UP000235965 UP000007755 UP000078200 UP000092462 UP000078541 UP000245037 UP000215335 UP000005205 UP000000311 UP000078492 UP000092461 UP000078540 UP000076502 UP000076858 UP000078542 UP000000304 UP000002358 UP000007819 UP000007798
Interpro
Gene 3D
ProteinModelPortal
A0A2H1V472
S4PTE8
H9IXF5
A0A2Z5UIW9
A0A2A4K697
A0A212FGZ3
+ More
A0A3S2M8A6 A0A194QKG5 A0A194RAW8 Q0IG24 A0A084VYH0 A0A1B3PD61 A0A182QPS2 A0A182H4C5 N6TZA5 J3JT85 Q7PWU7 A0A1S4GZE9 A0A182XDI6 A0A1B3PDI8 A0A182KNH0 A0A182VJW1 A0A182NUP0 A0A2M4ASN9 B4P156 A0A182MAC8 A0A0J9R038 A0A1A9YH44 A0A2M4AS86 Q8SX32 B4HWZ7 W5JVJ4 A0A182UHS3 A0A1B0C0R2 A0A1B0FCW3 A0A0T6ATR6 A0A1A9ZEY4 A0A1B3PDG5 A0A182I732 A0A1I8N6K0 B3N4Z5 A0A139WG38 B3MK00 A0A182VU60 A0A182Y972 T1DTM4 A0A2M4BSV7 A0A182P9Q1 A0A1A9WLY0 A0A182J705 A0A2M4BSV1 A0A182R4A0 B4KHQ5 A0A1W4VSY3 A0A182K391 B4LVB2 A0A3B0K6Q8 A0A2M3ZBC0 A0A2M3ZB12 A0A1J1IXV1 A0A182F1Z2 B4JQ09 A0A0M4EQJ1 A0A182SXK9 A0A1B6IYR1 B0WU19 A0A2J7R7Y1 A0A2J7R7W6 A0A2M4ASL5 F4WX56 A0A1B6DBT6 A0A1A9VI55 A0A1B0DD49 A0A195FAA6 A0A2P8XI50 A0A1B6KTC2 A0A1Y1MT54 A0A232FIH2 A0A158NCJ5 E2ALX0 A0A195EMT1 A0A1L8DM44 A0A1B0GL87 E9IM91 A0A195BWH1 A0A336LS43 A0A0P5CQZ0 A0A154P2Z9 A0A0N8CF82 A0A195CS44 B4QA90 A0A0P5FS48 K7IQH8 J9JST9 B4MUA4 A0A2S2QSN3 A0A0N8B5X4 A0A0P5IW72 A0A2S2PIV1 A0A0P5CXL8
A0A3S2M8A6 A0A194QKG5 A0A194RAW8 Q0IG24 A0A084VYH0 A0A1B3PD61 A0A182QPS2 A0A182H4C5 N6TZA5 J3JT85 Q7PWU7 A0A1S4GZE9 A0A182XDI6 A0A1B3PDI8 A0A182KNH0 A0A182VJW1 A0A182NUP0 A0A2M4ASN9 B4P156 A0A182MAC8 A0A0J9R038 A0A1A9YH44 A0A2M4AS86 Q8SX32 B4HWZ7 W5JVJ4 A0A182UHS3 A0A1B0C0R2 A0A1B0FCW3 A0A0T6ATR6 A0A1A9ZEY4 A0A1B3PDG5 A0A182I732 A0A1I8N6K0 B3N4Z5 A0A139WG38 B3MK00 A0A182VU60 A0A182Y972 T1DTM4 A0A2M4BSV7 A0A182P9Q1 A0A1A9WLY0 A0A182J705 A0A2M4BSV1 A0A182R4A0 B4KHQ5 A0A1W4VSY3 A0A182K391 B4LVB2 A0A3B0K6Q8 A0A2M3ZBC0 A0A2M3ZB12 A0A1J1IXV1 A0A182F1Z2 B4JQ09 A0A0M4EQJ1 A0A182SXK9 A0A1B6IYR1 B0WU19 A0A2J7R7Y1 A0A2J7R7W6 A0A2M4ASL5 F4WX56 A0A1B6DBT6 A0A1A9VI55 A0A1B0DD49 A0A195FAA6 A0A2P8XI50 A0A1B6KTC2 A0A1Y1MT54 A0A232FIH2 A0A158NCJ5 E2ALX0 A0A195EMT1 A0A1L8DM44 A0A1B0GL87 E9IM91 A0A195BWH1 A0A336LS43 A0A0P5CQZ0 A0A154P2Z9 A0A0N8CF82 A0A195CS44 B4QA90 A0A0P5FS48 K7IQH8 J9JST9 B4MUA4 A0A2S2QSN3 A0A0N8B5X4 A0A0P5IW72 A0A2S2PIV1 A0A0P5CXL8
PDB
5G02
E-value=4.02888e-56,
Score=551
Ontologies
GO
PANTHER
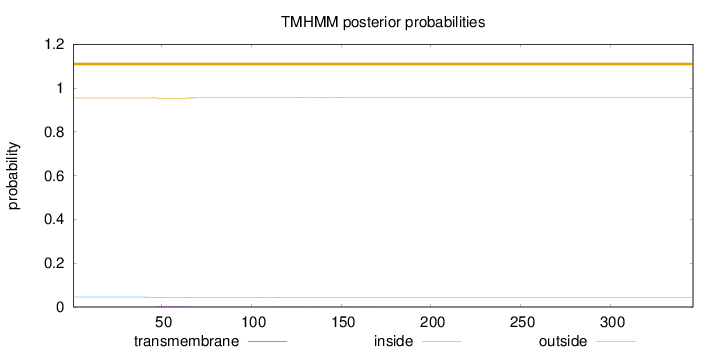
Topology
Length:
346
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0872699999999999
Exp number, first 60 AAs:
0.04461
Total prob of N-in:
0.04511
outside
1 - 346
Population Genetic Test Statistics
Pi
209.199433
Theta
165.903238
Tajima's D
1.116213
CLR
0.309705
CSRT
0.694115294235288
Interpretation
Uncertain
