Pre Gene Modal
BGIBMGA001886
Annotation
PREDICTED:_gamma-tubulin_complex_component_4_[Bombyx_mori]
Full name
Gamma-tubulin complex component
+ More
Guanylate cyclase
Gamma-tubulin complex component 4
Guanylate cyclase
Gamma-tubulin complex component 4
Alternative Name
Gamma-ring complex protein 76 kDa
Location in the cell
Nuclear Reliability : 2.356
Sequence
CDS
ATGATTCATGACGTACTATTAATTCTTTGGGATTCATCTGAAAATGCTATCGAAAAAAAAGAGAACCTTGCTTATGCGAATGAGGGTGATGTTGAAATCTATGGTAGATTAAGTGAGATAGCAAAGAGCCATCATAGAATCCAATCCTTTGTAGAAGAGACTGGATTCCAATCCCGAAAAGCTTTATCAGCAACAACACACAGTCACAGATATGGACTCTATACCCAAGCACTAAGTGAAGGCATCACTGAAGTCCTACTATCATATAAGAAAACACTTGTAGAAGTTGAATCTCTAGTAATAGGGAATCACAACTACACTTTATCATATGTCTATAGTTACATTGAGAAATATCAAGGTCTGTTCAATACATTGAACAGAATTATAACAACTATTAAAGAAAGGAGATTAATGGGCTGTCAGATATTGAGTTTAATTCATCAATATATTTTAAGTGGCAACAAGTCGATTCATGATGCCGTTTTGAAAATATTTACAAGTGTGAACAAAGTATTCATACATCAACTATGTACTTGGCTTCTTTATGGGGAATTAAAGGATGTGTATCAAGAGTTCTTCATATCAAAAAGAAAAGATGACTCGTCAGAAACTTTATTGTCAACACCATCTTCTTCTAATGTCTGTGACAAAACTCTTGTTTCTACATTGCAGGAGACATTAGGTTGTGGTTACACACTCAATCCAACACTGATACCACATTTCATGCCATTGTCATTAGCACAAGAAATTTTGTTTATCGGAAAAACTGTAATACTTTTCGATTTAGATCCAAAAAAAGTAAAAAAACACAGTGGCTTCGCTTCAAGCAGACCGATGTTGCCTTTGAAAAAGTATGGAATTGAGTCTAGAAGGTTTACAATTTGGGAGGACAAAGAATCTGAATTTCATGAGAAACTACAAAAACTTAAAGATCCAGATAATTTTGAAGTTCATAATCTGAGAAGTGTTATAAGGGAGATTAAAGAATGTGTAACTAAGCATTTGTGGACAGTGGCAGTCGAAGAAGCCCAATTGATACATGAGTTGAAGCTAATGAAAGATTTCTATCTACTTGGCCGGGGCGAATTGTTTCTGGAATTATTAAGACTCACAGCACATATGCTGGACAAACCGACGATTCGCACGTCAAGTAGAGACATGAACCAGGCTTTTCAAATGGCGGCACGCTCCGTTTTCCTAAGTAATAATACTGATATAGAAAAGTTTAGTTTCGAGCTGCCTTATGTTAAGCCGAACATATCAGGGAATTCGTCGATTGACGAAGACTACAGTACCGTTGCAGATGGATGGTCGAGTATAATCTTGAAATATGATTTCAAATGGCCGTTACATTTATTATTTACACCGGAAGTACTTGCCCGTTATAATGATATGTTTCGACTATTGCTCCGGCTAAAGAAAACCCAACACGATCTGCATTCGCTTTGGAAAACGTACAAACAGTCGGCGAGTTTCGCGATGTCCCAACTACACAACAAACTGATGTTTCTAATGGACAACCTCCAACACTACATGCAAGCAGACGTATTAGAAACCAATTTCTCTCGTCTGATGGACGCTGTCAACAAGACAAACGACTTTGAGAAACTCAAGAAGGCTCACGCCACGTTCCTGGCTGATATATTGAGTCAGTCATTTTTGACTGTGACTTTGTCGTCAGATAGCGATTGTTCTGGAGATCCCGCCGATACATTAAATAACCCAGTGTTCTGTAACATTATGGAACTATTGAAATTGTGTCATTCATTCTGCAGTATGAGTGAGATAGCAAACGATAGGGAAGCTGAAGATTATTACATAAAAGGATATTCTGAAAGGTTCAACAAATTAGTGAAGCAACTAATGCAACTTCTAGTTTCGCTACGAGATCGTCCGTGCGGCGTTTACTTAGCGAGACTTCTGATGAGACTCGACTACAACCGGTGGCTCAGTAGAGAGGCGCAGTTAACGCAATCTGTAACGAACATGTTAAGATACTGA
Protein
MIHDVLLILWDSSENAIEKKENLAYANEGDVEIYGRLSEIAKSHHRIQSFVEETGFQSRKALSATTHSHRYGLYTQALSEGITEVLLSYKKTLVEVESLVIGNHNYTLSYVYSYIEKYQGLFNTLNRIITTIKERRLMGCQILSLIHQYILSGNKSIHDAVLKIFTSVNKVFIHQLCTWLLYGELKDVYQEFFISKRKDDSSETLLSTPSSSNVCDKTLVSTLQETLGCGYTLNPTLIPHFMPLSLAQEILFIGKTVILFDLDPKKVKKHSGFASSRPMLPLKKYGIESRRFTIWEDKESEFHEKLQKLKDPDNFEVHNLRSVIREIKECVTKHLWTVAVEEAQLIHELKLMKDFYLLGRGELFLELLRLTAHMLDKPTIRTSSRDMNQAFQMAARSVFLSNNTDIEKFSFELPYVKPNISGNSSIDEDYSTVADGWSSIILKYDFKWPLHLLFTPEVLARYNDMFRLLLRLKKTQHDLHSLWKTYKQSASFAMSQLHNKLMFLMDNLQHYMQADVLETNFSRLMDAVNKTNDFEKLKKAHATFLADILSQSFLTVTLSSDSDCSGDPADTLNNPVFCNIMELLKLCHSFCSMSEIANDREAEDYYIKGYSERFNKLVKQLMQLLVSLRDRPCGVYLARLLMRLDYNRWLSREAQLTQSVTNMLRY
Summary
Description
Gamma-tubulin complex is necessary for microtubule nucleation at the centrosome.
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Subunit
Gamma-tubulin complex is composed of gamma-tubulin, TUBGCP2, TUBGCP3, TUBGCP4, TUBGCP5 and TUBGCP6. Interacts with NINL. Interacts with ATF5; the ATF5:PCNT:polyglutamylated tubulin (PGT) tripartite unites the mother centriole and the pericentriolar material (PCM) in the centrosome (PubMed:26213385).
Similarity
Belongs to the TUBGCP family.
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Keywords
3D-structure
Alternative splicing
Complete proteome
Cytoplasm
Cytoskeleton
Direct protein sequencing
Mental retardation
Microtubule
Primary microcephaly
Reference proteome
Feature
chain Gamma-tubulin complex component
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2A4K4L0
A0A2H1V5U6
A0A212FGZ7
A0A194QEP6
H9IXA3
A0A194RA79
+ More
A0A2Z5U7B5 A0A154P4F8 E2AHZ6 K7J485 A0A1Q3F4W2 A0A1Q3F4S0 A0A2A3EMN2 A0A088AL65 B0WU20 A0A310SEX1 A0A026WTB7 A0A2J7QYH5 A0A158NL53 A0A0M8ZUL3 A0A0C9R8R5 A0A067RAM5 Q0IG23 E2B6I6 A0A195C034 A0A0L7R764 A0A182K390 A0A151JSQ1 A0A195F1M9 F4WFW3 A0A151XBJ8 A0A182IW86 A0A084VYH1 A0A182VFJ6 W5JU71 A0A182NUP1 A0A182Y971 Q7PWU6 A0A2C9GRZ4 A0A182KNH6 A0A336L0T3 A0A336MBV4 A0A182R499 A0A1J1I5S3 A0A182MP26 A0A182XDI7 A0A182F1Z3 A0A1S4F4Z4 A0A0C9QLV5 A0A182VU61 A0A0J7L5U5 A0A1B0FCW2 A0A182P9Q0 A0A182H916 A0A182TPB3 A0A195BH14 A0A1A9VI50 A0A1A9YH42 A0A1A9ZEY2 A0A3Q3X7X3 A0A3P8VJ44 A0A3Q0S2N5 A0A3Q2PPM5 A0A146V2N1 B4KHQ7 A0A3P9PUZ3 A0A3B4CXV7 A0A087XQL3 A0A3B3TMV5 A0A3B3WMY5 A0A3B4YQQ2 A0A3B4V6S8 I3JIZ7 H2UBS6 A0A3Q1J2N3 A0A1A8HAU1 A0A232F3N5 A0A3Q2WKJ7 I3JIZ6 A0A1V4JDX7 A0A3Q4H2K7 A0A1A8K099 A0A1A8QIU2 W5NA20 A0A1W5B1M1 M4AUZ7 E1BQM2 A0A1A8DHC5 A0A1A7YDD3 A0A2I4CA55 A0A3P9CG09 Q9UGJ1-2 K7C990 A0A2I2Y2W4 A0A3Q3FQ69
A0A2Z5U7B5 A0A154P4F8 E2AHZ6 K7J485 A0A1Q3F4W2 A0A1Q3F4S0 A0A2A3EMN2 A0A088AL65 B0WU20 A0A310SEX1 A0A026WTB7 A0A2J7QYH5 A0A158NL53 A0A0M8ZUL3 A0A0C9R8R5 A0A067RAM5 Q0IG23 E2B6I6 A0A195C034 A0A0L7R764 A0A182K390 A0A151JSQ1 A0A195F1M9 F4WFW3 A0A151XBJ8 A0A182IW86 A0A084VYH1 A0A182VFJ6 W5JU71 A0A182NUP1 A0A182Y971 Q7PWU6 A0A2C9GRZ4 A0A182KNH6 A0A336L0T3 A0A336MBV4 A0A182R499 A0A1J1I5S3 A0A182MP26 A0A182XDI7 A0A182F1Z3 A0A1S4F4Z4 A0A0C9QLV5 A0A182VU61 A0A0J7L5U5 A0A1B0FCW2 A0A182P9Q0 A0A182H916 A0A182TPB3 A0A195BH14 A0A1A9VI50 A0A1A9YH42 A0A1A9ZEY2 A0A3Q3X7X3 A0A3P8VJ44 A0A3Q0S2N5 A0A3Q2PPM5 A0A146V2N1 B4KHQ7 A0A3P9PUZ3 A0A3B4CXV7 A0A087XQL3 A0A3B3TMV5 A0A3B3WMY5 A0A3B4YQQ2 A0A3B4V6S8 I3JIZ7 H2UBS6 A0A3Q1J2N3 A0A1A8HAU1 A0A232F3N5 A0A3Q2WKJ7 I3JIZ6 A0A1V4JDX7 A0A3Q4H2K7 A0A1A8K099 A0A1A8QIU2 W5NA20 A0A1W5B1M1 M4AUZ7 E1BQM2 A0A1A8DHC5 A0A1A7YDD3 A0A2I4CA55 A0A3P9CG09 Q9UGJ1-2 K7C990 A0A2I2Y2W4 A0A3Q3FQ69
EC Number
4.6.1.2
Pubmed
22118469
26354079
19121390
20798317
20075255
24508170
+ More
30249741 21347285 24845553 17510324 21719571 24438588 20920257 23761445 25244985 12364791 14747013 17210077 20966253 26483478 24487278 17994087 25186727 21551351 28648823 23542700 15592404 10562286 14702039 16572171 15489334 12852856 21269460 25817018 26213385 22398555
30249741 21347285 24845553 17510324 21719571 24438588 20920257 23761445 25244985 12364791 14747013 17210077 20966253 26483478 24487278 17994087 25186727 21551351 28648823 23542700 15592404 10562286 14702039 16572171 15489334 12852856 21269460 25817018 26213385 22398555
EMBL
NWSH01000136
PCG79191.1
ODYU01000592
SOQ35644.1
AGBW02008579
OWR53004.1
+ More
KQ459053 KPJ03952.1 BABH01018072 BABH01018073 BABH01018074 BABH01018075 KQ460436 KPJ14748.1 AP017509 BBB06823.1 KQ434809 KZC06733.1 GL439621 EFN66904.1 AAZX01003261 GFDL01012482 JAV22563.1 GFDL01012480 JAV22565.1 KZ288209 PBC32988.1 DS232099 EDS34736.1 KQ765447 OAD53938.1 KK107111 QOIP01000011 EZA58911.1 RLU16553.1 NEVH01009089 PNF33641.1 ADTU01019255 ADTU01019256 KQ435843 KOX71287.1 GBYB01004531 GBYB01004532 GBYB01004533 GBYB01004535 JAG74298.1 JAG74299.1 JAG74300.1 JAG74302.1 KK852779 KDR16711.1 CH477274 EAT45190.1 GL445975 EFN88678.1 KQ978501 KYM93518.1 KQ414642 KOC66715.1 KQ978530 KYN30314.1 KQ981866 KYN34077.1 GL888128 EGI66730.1 KQ982320 KYQ57727.1 ATLV01018366 KE525231 KFB43015.1 ADMH02000358 ETN66843.1 AAAB01008984 EAA14809.4 APCN01003350 UFQS01001202 UFQT01001202 SSX09724.1 SSX29491.1 UFQT01000665 SSX26293.1 CVRI01000040 CRK94932.1 AXCM01000043 GBYB01004534 JAG74301.1 LBMM01000598 KMQ97986.1 CCAG010017495 JXUM01030305 KQ560880 KXJ80500.1 KQ976488 KYM83518.1 GCES01074661 JAR11662.1 CH933807 EDW13344.1 AYCK01027203 AERX01043080 AERX01043081 AERX01043082 HAEB01007560 HAEC01012386 SBQ80603.1 NNAY01001043 OXU25374.1 LSYS01007908 OPJ70225.1 HAED01009278 HAEE01005622 SBR25642.1 HAEF01007349 HAEG01012832 SBR93362.1 AHAT01024787 AADN05000047 HAEA01005260 SBQ33740.1 HADX01006041 SBP28273.1 AJ249677 AK001639 AK027703 AC018924 CH471125 BC009870 BC012801 GABC01008888 GABF01002251 GABD01010221 NBAG03000046 JAA02450.1 JAA19894.1 JAA22879.1 PNI94641.1 CABD030095860 CABD030095861
KQ459053 KPJ03952.1 BABH01018072 BABH01018073 BABH01018074 BABH01018075 KQ460436 KPJ14748.1 AP017509 BBB06823.1 KQ434809 KZC06733.1 GL439621 EFN66904.1 AAZX01003261 GFDL01012482 JAV22563.1 GFDL01012480 JAV22565.1 KZ288209 PBC32988.1 DS232099 EDS34736.1 KQ765447 OAD53938.1 KK107111 QOIP01000011 EZA58911.1 RLU16553.1 NEVH01009089 PNF33641.1 ADTU01019255 ADTU01019256 KQ435843 KOX71287.1 GBYB01004531 GBYB01004532 GBYB01004533 GBYB01004535 JAG74298.1 JAG74299.1 JAG74300.1 JAG74302.1 KK852779 KDR16711.1 CH477274 EAT45190.1 GL445975 EFN88678.1 KQ978501 KYM93518.1 KQ414642 KOC66715.1 KQ978530 KYN30314.1 KQ981866 KYN34077.1 GL888128 EGI66730.1 KQ982320 KYQ57727.1 ATLV01018366 KE525231 KFB43015.1 ADMH02000358 ETN66843.1 AAAB01008984 EAA14809.4 APCN01003350 UFQS01001202 UFQT01001202 SSX09724.1 SSX29491.1 UFQT01000665 SSX26293.1 CVRI01000040 CRK94932.1 AXCM01000043 GBYB01004534 JAG74301.1 LBMM01000598 KMQ97986.1 CCAG010017495 JXUM01030305 KQ560880 KXJ80500.1 KQ976488 KYM83518.1 GCES01074661 JAR11662.1 CH933807 EDW13344.1 AYCK01027203 AERX01043080 AERX01043081 AERX01043082 HAEB01007560 HAEC01012386 SBQ80603.1 NNAY01001043 OXU25374.1 LSYS01007908 OPJ70225.1 HAED01009278 HAEE01005622 SBR25642.1 HAEF01007349 HAEG01012832 SBR93362.1 AHAT01024787 AADN05000047 HAEA01005260 SBQ33740.1 HADX01006041 SBP28273.1 AJ249677 AK001639 AK027703 AC018924 CH471125 BC009870 BC012801 GABC01008888 GABF01002251 GABD01010221 NBAG03000046 JAA02450.1 JAA19894.1 JAA22879.1 PNI94641.1 CABD030095860 CABD030095861
Proteomes
UP000218220
UP000007151
UP000053268
UP000005204
UP000053240
UP000076502
+ More
UP000000311 UP000002358 UP000242457 UP000005203 UP000002320 UP000053097 UP000279307 UP000235965 UP000005205 UP000053105 UP000027135 UP000008820 UP000008237 UP000078542 UP000053825 UP000075881 UP000078492 UP000078541 UP000007755 UP000075809 UP000075880 UP000030765 UP000075903 UP000000673 UP000075884 UP000076408 UP000007062 UP000075840 UP000075882 UP000075900 UP000183832 UP000075883 UP000076407 UP000069272 UP000075920 UP000036403 UP000092444 UP000075885 UP000069940 UP000249989 UP000075902 UP000078540 UP000078200 UP000092443 UP000092445 UP000261620 UP000265120 UP000261340 UP000265000 UP000009192 UP000242638 UP000261440 UP000028760 UP000261500 UP000261480 UP000261360 UP000261420 UP000005207 UP000005226 UP000265040 UP000215335 UP000264840 UP000190648 UP000261580 UP000018468 UP000192224 UP000002852 UP000000539 UP000192220 UP000265160 UP000005640 UP000001519 UP000261660
UP000000311 UP000002358 UP000242457 UP000005203 UP000002320 UP000053097 UP000279307 UP000235965 UP000005205 UP000053105 UP000027135 UP000008820 UP000008237 UP000078542 UP000053825 UP000075881 UP000078492 UP000078541 UP000007755 UP000075809 UP000075880 UP000030765 UP000075903 UP000000673 UP000075884 UP000076408 UP000007062 UP000075840 UP000075882 UP000075900 UP000183832 UP000075883 UP000076407 UP000069272 UP000075920 UP000036403 UP000092444 UP000075885 UP000069940 UP000249989 UP000075902 UP000078540 UP000078200 UP000092443 UP000092445 UP000261620 UP000265120 UP000261340 UP000265000 UP000009192 UP000242638 UP000261440 UP000028760 UP000261500 UP000261480 UP000261360 UP000261420 UP000005207 UP000005226 UP000265040 UP000215335 UP000264840 UP000190648 UP000261580 UP000018468 UP000192224 UP000002852 UP000000539 UP000192220 UP000265160 UP000005640 UP000001519 UP000261660
Pfam
Interpro
IPR040457
GCP_C
+ More
IPR007259 GCP
IPR042241 GCP_C_sf
IPR041470 GCP_N
IPR011009 Kinase-like_dom_sf
IPR018297 A/G_cyclase_CS
IPR000719 Prot_kinase_dom
IPR029787 Nucleotide_cyclase
IPR001828 ANF_lig-bd_rcpt
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001054 A/G_cyclase
IPR028082 Peripla_BP_I
IPR029752 D-isomer_DH_CS1
IPR029753 D-isomer_DH_CS
IPR006139 D-isomer_2_OHA_DH_cat_dom
IPR006140 D-isomer_DH_NAD-bd
IPR036291 NAD(P)-bd_dom_sf
IPR004117 7tm6_olfct_rcpt
IPR018247 EF_Hand_1_Ca_BS
IPR007259 GCP
IPR042241 GCP_C_sf
IPR041470 GCP_N
IPR011009 Kinase-like_dom_sf
IPR018297 A/G_cyclase_CS
IPR000719 Prot_kinase_dom
IPR029787 Nucleotide_cyclase
IPR001828 ANF_lig-bd_rcpt
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001054 A/G_cyclase
IPR028082 Peripla_BP_I
IPR029752 D-isomer_DH_CS1
IPR029753 D-isomer_DH_CS
IPR006139 D-isomer_2_OHA_DH_cat_dom
IPR006140 D-isomer_DH_NAD-bd
IPR036291 NAD(P)-bd_dom_sf
IPR004117 7tm6_olfct_rcpt
IPR018247 EF_Hand_1_Ca_BS
Gene 3D
ProteinModelPortal
A0A2A4K4L0
A0A2H1V5U6
A0A212FGZ7
A0A194QEP6
H9IXA3
A0A194RA79
+ More
A0A2Z5U7B5 A0A154P4F8 E2AHZ6 K7J485 A0A1Q3F4W2 A0A1Q3F4S0 A0A2A3EMN2 A0A088AL65 B0WU20 A0A310SEX1 A0A026WTB7 A0A2J7QYH5 A0A158NL53 A0A0M8ZUL3 A0A0C9R8R5 A0A067RAM5 Q0IG23 E2B6I6 A0A195C034 A0A0L7R764 A0A182K390 A0A151JSQ1 A0A195F1M9 F4WFW3 A0A151XBJ8 A0A182IW86 A0A084VYH1 A0A182VFJ6 W5JU71 A0A182NUP1 A0A182Y971 Q7PWU6 A0A2C9GRZ4 A0A182KNH6 A0A336L0T3 A0A336MBV4 A0A182R499 A0A1J1I5S3 A0A182MP26 A0A182XDI7 A0A182F1Z3 A0A1S4F4Z4 A0A0C9QLV5 A0A182VU61 A0A0J7L5U5 A0A1B0FCW2 A0A182P9Q0 A0A182H916 A0A182TPB3 A0A195BH14 A0A1A9VI50 A0A1A9YH42 A0A1A9ZEY2 A0A3Q3X7X3 A0A3P8VJ44 A0A3Q0S2N5 A0A3Q2PPM5 A0A146V2N1 B4KHQ7 A0A3P9PUZ3 A0A3B4CXV7 A0A087XQL3 A0A3B3TMV5 A0A3B3WMY5 A0A3B4YQQ2 A0A3B4V6S8 I3JIZ7 H2UBS6 A0A3Q1J2N3 A0A1A8HAU1 A0A232F3N5 A0A3Q2WKJ7 I3JIZ6 A0A1V4JDX7 A0A3Q4H2K7 A0A1A8K099 A0A1A8QIU2 W5NA20 A0A1W5B1M1 M4AUZ7 E1BQM2 A0A1A8DHC5 A0A1A7YDD3 A0A2I4CA55 A0A3P9CG09 Q9UGJ1-2 K7C990 A0A2I2Y2W4 A0A3Q3FQ69
A0A2Z5U7B5 A0A154P4F8 E2AHZ6 K7J485 A0A1Q3F4W2 A0A1Q3F4S0 A0A2A3EMN2 A0A088AL65 B0WU20 A0A310SEX1 A0A026WTB7 A0A2J7QYH5 A0A158NL53 A0A0M8ZUL3 A0A0C9R8R5 A0A067RAM5 Q0IG23 E2B6I6 A0A195C034 A0A0L7R764 A0A182K390 A0A151JSQ1 A0A195F1M9 F4WFW3 A0A151XBJ8 A0A182IW86 A0A084VYH1 A0A182VFJ6 W5JU71 A0A182NUP1 A0A182Y971 Q7PWU6 A0A2C9GRZ4 A0A182KNH6 A0A336L0T3 A0A336MBV4 A0A182R499 A0A1J1I5S3 A0A182MP26 A0A182XDI7 A0A182F1Z3 A0A1S4F4Z4 A0A0C9QLV5 A0A182VU61 A0A0J7L5U5 A0A1B0FCW2 A0A182P9Q0 A0A182H916 A0A182TPB3 A0A195BH14 A0A1A9VI50 A0A1A9YH42 A0A1A9ZEY2 A0A3Q3X7X3 A0A3P8VJ44 A0A3Q0S2N5 A0A3Q2PPM5 A0A146V2N1 B4KHQ7 A0A3P9PUZ3 A0A3B4CXV7 A0A087XQL3 A0A3B3TMV5 A0A3B3WMY5 A0A3B4YQQ2 A0A3B4V6S8 I3JIZ7 H2UBS6 A0A3Q1J2N3 A0A1A8HAU1 A0A232F3N5 A0A3Q2WKJ7 I3JIZ6 A0A1V4JDX7 A0A3Q4H2K7 A0A1A8K099 A0A1A8QIU2 W5NA20 A0A1W5B1M1 M4AUZ7 E1BQM2 A0A1A8DHC5 A0A1A7YDD3 A0A2I4CA55 A0A3P9CG09 Q9UGJ1-2 K7C990 A0A2I2Y2W4 A0A3Q3FQ69
PDB
3RIP
E-value=1.49585e-76,
Score=730
Ontologies
KEGG
GO
GO:0000922
GO:0007020
GO:0005815
GO:0005737
GO:0043015
GO:0005874
GO:0051225
GO:0005813
GO:0000923
GO:0000930
GO:0031122
GO:0000278
GO:0051321
GO:0051415
GO:0004383
GO:0035556
GO:0005524
GO:0004672
GO:0016021
GO:0016616
GO:0051287
GO:0004984
GO:0005549
GO:0007112
GO:0007058
GO:0045450
GO:0060221
GO:0042670
GO:0055037
GO:0008274
GO:0005200
GO:0005829
GO:0015630
GO:0016020
GO:0065003
GO:0000226
GO:0008152
GO:0016740
GO:0003707
GO:0006418
GO:0003824
PANTHER
Topology
Subcellular location
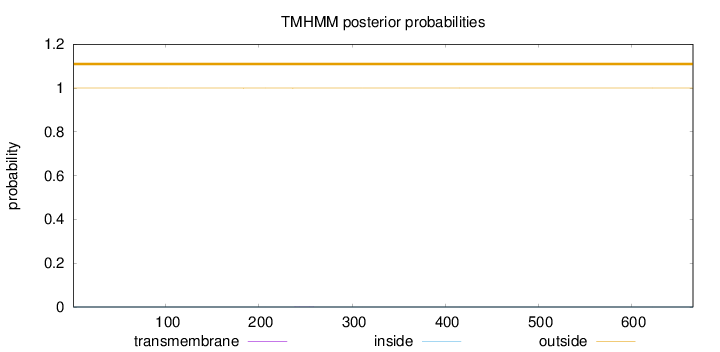
Length:
666
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00975999999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00029
outside
1 - 666
Population Genetic Test Statistics
Pi
182.768581
Theta
170.598505
Tajima's D
0.348446
CLR
0.031048
CSRT
0.476126193690316
Interpretation
Uncertain
