Gene
KWMTBOMO11428
Pre Gene Modal
BGIBMGA001939
Annotation
PREDICTED:_uncharacterized_protein_LOC101746965_[Bombyx_mori]
Full name
XK-related protein
Location in the cell
PlasmaMembrane Reliability : 3.352
Sequence
CDS
ATGGCAGAGGTTTGCAAAACTAGTAGTGGTACTACTGAAAATAAATTGAGTAATACTGATGTACACCAAGATGAAAACTTTTGTTTATTTAATGTATTATGCGTTTTCATATCGTTGGTTTCTATATTAGCTGATGTAACCACAGATATAATTGTATTTGCGGAATATTTTAAAAACGATCGTTTAGAATGGGCAGCAGGCACAATCATTTTAATAATAATTCCAAACATTATAATCAATGTGTTTTCTTTGAGATGGCTTATTACTGATAATAAATCCAATGTAACTCATTGGATTAGCCACATTTTCCTGGTTGGATTGTTAGAAAGATATTTCATATTCATACGCAAAGTGTTTGGATGTAAAAATTTAGAAGCTATGAAAACAAACAAACTGATATCCCAAAGGAATGATCTAAGTTTGTTGCATTTTATTTACATATTTACTGGAACGGCCCCACAAATTATACTACAGCTTTATGCCATTGTTGTTCTGGATGAAAACTACTATACTAAAGTATTTTCATTGAGTGCCTCCATGTCTTCAGTTGTATGGGGGTGCTCAACATACATATACAATGAGGCAGCTTTCCTGAAGTTACAGTTTGAATGGAAATCATTAGGATTGAGGCTTACGTGGTTCTTCGGTATGTTAATAAGCAGAATTTCGGGAATTTTGCTATTTACTATAGTATTTGGAGTTTGGACATTGTTACCACTAAGCTTACACTGGACAGCTATGATATTCTGGATTATTATTCAAAAAACCAGCTTTTGTTCTAATAAATTAGAGGAAACATTTTATAATGTAGTAATGGGATTTGTTTATTGTTTTTGCTACGTTAATTTACACGAGGGCCAAACTAAATATAGACTATTGACGTTTTACACATTAATGGTAACTCAAAATTTCGGTAGTCTCCTATTGTACTTGCTTATTTCGCCGCACGACAAGCAAAGGAAGTTATGGTCTGTGATAGGAATCGTTTGCATTATTTTTGGTGTATTTATAGGTATTTGTGCTATGATATTTTACTATAGATTTTATCATCCGAAAGGACCACTATTGTGGAAAAGAAAACAAAGTGATATAGAATTAAATAATAAGGCTGTGATACAAAACCCCGGAACCGAAGAAAATAAGACGGTTGTCAATTTAAATCCCATTAGGTCATTTAAACATTCTCTGCATCGTAATCAAAATGGAGCCTCTTACAGCATTGAATCAACTTTAGAACAACATCCCAAAAAAGACATCGAAAACGCTAATAAGAATTTTTTGCTTGATCAATGGATTGCTAAAATTGGCGTACCGACGTTATCGTCCGGCTCATTTGGAAACAATGGATGCGATGCTTCAGGAATCGAAATTTATAGTGTTGAAAATACACCACAAACTCTCAAACAGGATAATCAAGGGCAAAATATTTCAAATAAAATTAACTGTAAACACAAAAAAATTCTAATATGTTCACCAACGGAACTGACGCTTAAACTATCGGCTGGAGAAAATATTGAGAGTAATATAGATGAAATAACATCGAGCAATCTGAATTCAAATAAAAGACGCGCCATTTGTTCCTCTGATGATCTTAGCGATGATTTAACCAATATGAATTTCGACGTTTGTGCCATAGTCAATGAAGATATTATGAAGATACTCAAAAACGACCAATGCTCAGAAATAAGTCACGATACCGATAGCTCGAATAAGAAAACTTCATCAGATTCCATTGATATGGATTTAGATTTGAGTTTCAGCGACATCAATAGTTCTGATATCAACACGTTTAACGAAAACATAGTCCAGAAATTGTGTTTAAGTGCTCTCAAGAATATAAAAGTAGGGAATCAGGATGCAGATATTGAAAACATACAAAGGATCGCATTGGACATTCTAAAAGAAATGTACGCTAAAAAGGGTAAGAAGAGCAATGCGAATCCAGATTTCTTTACAAAATCAAGAGAGTTAGATACGCCAACGGAAATATTGTCCGTACACGATTATGAGAATATATGTGCGGTAAATATTGCTCGTGAAGCGTGGGGGCTGCGATCTTGGAAGGGGTACTGCGATATCGAAAACTGGCGTCACGACGGAAGCGTTGTCAGAGATAGAAGACGCGACACATTAACTTCGGCCAGTTCAGAAATTAGTAGTTGCATCTCTCAGGAATTGAAAAATGCGATTTTAGCGGCACCACCTTTCCCGAAGAAAACACATACATACTCGAATGTATTCGTTAAGTTTGACAAAGGCAGTCAAGATGACTACGCAAACTCTACAATATGTACCGATGACTCATTTAGCGTAGTTGATGCTAATAAAGATGCAATTCATTTAGAACCAATTATGGAGGAATTGGATGACTTGACAGACATGGATATTACGAACAAGAAACGGTTAAATTCAGAGAGCTCTTTAGTTGCAACTATCGATGAGATTAGGAAATCTACTGTTACAAATTCTCCAAGAAATCTATATCATAGAGGAGACCATTTGTGGGATTCACCTCAACCGACTGCTTCAAACTTAGACCCGATTTTATCCGAAGCATTATGGAAAGAACCTTCGAAATTCCATCCCAGTAATATTATGGAATCATTGGCTGAAAAGGAAAACGATAAAAATATATCACAAAATCCCAGTTATGACACAAATAGAAAAACTTTAAACGTGACAGATACAAACAATTCAAAACACGATACATTAAGAATGAAAATAGAAAATAATTTCAATAATCACTCAACAAACTACACTCGTAGTAAAATGAAAAACTTGAAGACGCTACCTGATCCTGATGTATCTAATAAGAATATAGACTTGCTGTGGCAGTTGGTTTCAGGAAATGCCACAACAGCGAATGGTTCTAACGATTATGTATCAATGAATCGTATGTCAACATCTCTGCAATCTTTAATACAAAAAGACGTTTCGGCATCAGAATCAAATAAAAGCTCTTCGCCAAAACTTTCAAATATAAAAAAACAAACAAAAACCAAACCTCGTCGTAAGTTCTCTATACTAAGAGAAAAATTTGAAATGAAAACACATTTAAACTCTGATGAGGAAAATTTGTGTAATACTTCTGAAAGTACATCAAAAACACTTGATGTCATGTCAAGCAAAAATAATTATAGATCGAGTGATAAGGTCACAACCATAGAGAAAGAATCAAGTACTTATTGGCCAGGTACCTCTCCTACAGGTAGTACAAGCTCTAATAATTCCAACTGTGCAAAGGAGAAAAGATCACTATTTATGAAACAGGTATTAAGTCCATCTAAATCTAATACAAAGATGAAACACAAGGGCTGA
Protein
MAEVCKTSSGTTENKLSNTDVHQDENFCLFNVLCVFISLVSILADVTTDIIVFAEYFKNDRLEWAAGTIILIIIPNIIINVFSLRWLITDNKSNVTHWISHIFLVGLLERYFIFIRKVFGCKNLEAMKTNKLISQRNDLSLLHFIYIFTGTAPQIILQLYAIVVLDENYYTKVFSLSASMSSVVWGCSTYIYNEAAFLKLQFEWKSLGLRLTWFFGMLISRISGILLFTIVFGVWTLLPLSLHWTAMIFWIIIQKTSFCSNKLEETFYNVVMGFVYCFCYVNLHEGQTKYRLLTFYTLMVTQNFGSLLLYLLISPHDKQRKLWSVIGIVCIIFGVFIGICAMIFYYRFYHPKGPLLWKRKQSDIELNNKAVIQNPGTEENKTVVNLNPIRSFKHSLHRNQNGASYSIESTLEQHPKKDIENANKNFLLDQWIAKIGVPTLSSGSFGNNGCDASGIEIYSVENTPQTLKQDNQGQNISNKINCKHKKILICSPTELTLKLSAGENIESNIDEITSSNLNSNKRRAICSSDDLSDDLTNMNFDVCAIVNEDIMKILKNDQCSEISHDTDSSNKKTSSDSIDMDLDLSFSDINSSDINTFNENIVQKLCLSALKNIKVGNQDADIENIQRIALDILKEMYAKKGKKSNANPDFFTKSRELDTPTEILSVHDYENICAVNIAREAWGLRSWKGYCDIENWRHDGSVVRDRRRDTLTSASSEISSCISQELKNAILAAPPFPKKTHTYSNVFVKFDKGSQDDYANSTICTDDSFSVVDANKDAIHLEPIMEELDDLTDMDITNKKRLNSESSLVATIDEIRKSTVTNSPRNLYHRGDHLWDSPQPTASNLDPILSEALWKEPSKFHPSNIMESLAEKENDKNISQNPSYDTNRKTLNVTDTNNSKHDTLRMKIENNFNNHSTNYTRSKMKNLKTLPDPDVSNKNIDLLWQLVSGNATTANGSNDYVSMNRMSTSLQSLIQKDVSASESNKSSSPKLSNIKKQTKTKPRRKFSILREKFEMKTHLNSDEENLCNTSESTSKTLDVMSSKNNYRSSDKVTTIEKESSTYWPGTSPTGSTSSNNSNCAKEKRSLFMKQVLSPSKSNTKMKHKG
Summary
Similarity
Belongs to the XK family.
Uniprot
EMBL
Proteomes
Pfam
PF09815 XK-related
CDD
ProteinModelPortal
Ontologies
GO
Topology
Subcellular location
Membrane
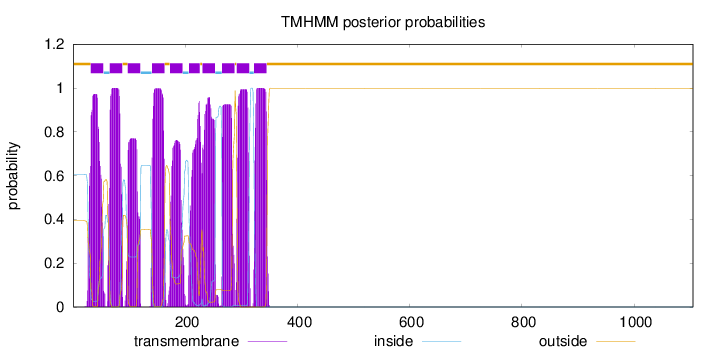
Length:
1105
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
199.10988
Exp number, first 60 AAs:
21.23315
Total prob of N-in:
0.60571
POSSIBLE N-term signal
sequence
outside
1 - 31
TMhelix
32 - 54
inside
55 - 65
TMhelix
66 - 88
outside
89 - 97
TMhelix
98 - 120
inside
121 - 140
TMhelix
141 - 163
outside
164 - 172
TMhelix
173 - 195
inside
196 - 206
TMhelix
207 - 226
outside
227 - 230
TMhelix
231 - 253
inside
254 - 265
TMhelix
266 - 288
outside
289 - 291
TMhelix
292 - 314
inside
315 - 322
TMhelix
323 - 345
outside
346 - 1105
Population Genetic Test Statistics
Pi
207.828844
Theta
186.014506
Tajima's D
0.251626
CLR
0.100601
CSRT
0.443477826108695
Interpretation
Uncertain
