Gene
KWMTBOMO11427
Pre Gene Modal
BGIBMGA001885
Annotation
Zyxin/trip6_[Operophtera_brumata]
Location in the cell
Extracellular Reliability : 2.202 Nuclear Reliability : 2.44
Sequence
CDS
ATGCACATTCAATTACCACCCCCTCCACCACCACCACCATTAACTACACACACAACATTGCAGGCTCCTCTAGGAACACACAATCTACAATATATGACACTGCCATTACCTAATCCTTCTTCAGGAGTCCACCAGGAATATATGATGCCAGTATTGTCCAAGTCTCCTGTAACTACAAACAGTGAGGAGTATATGCCACCCCCATCACCAAATAATTTCCCTGAATACAACATGTCACAAGCTCCAACATATGAATCATTCTATGAACCAATCAGTCCACATCCCTCCAGTAAGACAGCAATGCAAGAAAATAATTTAATTACAAAGAAAGAAGCTTTATCTAAACGATCACCATTGCCCAAGGAACAAGAAGTAGATGCTTTGACAAATCTGCTGGTACAATCAATTACAGACAGTCAAGATTTAGATGTCTTTGGAATATGTGTGAAGTGTGGTGAACGAATTTCTGGAGAAAACGCAGGCTGTACAGCTATGGGAAACACTTATCATGTTCACTGTTTCACTTGTCAACGTTGTAATGTAAATCTCCAAGGAAAACCATTCTATGATGTTGAGAATGAACCGTATTGTGAAGCTGATTATTATGACACATTAGAAAAGTGCTGTGTTTGCCGAAAAATAATTTTAGATCGTATTCTACGTGCTACTGGAAAACCTTATCATCCAACTTGTTTTTCTTGTGTAGAGTGTGGTAAAAGTTTAGATGGGATACCGTTTACAGTTGATGCCATGAACCAAATCCATTGTATTGATGACTTCCATAAGAAATTTGCACCACGTTGCTGTGTATGTGAACTACCAATTATGCCAGAAGAAGGTCAAGAGGAAACTGTGAGAGTTGTTGCCTTGGACCGTAGTTTCCATGTTAAGTGTTACAGGTGTGAAGATTGTGGCTTACTGTTGAGTTCAGAAGCAGAAGGTCGAGGTTGTTATCCACTTGACGATCATATACTTTGTAAAACTTGCAATGCACGTCGTGTTCGTCTTTTGACTAATGTTATGACTACAGACTTATGA
Protein
MHIQLPPPPPPPPLTTHTTLQAPLGTHNLQYMTLPLPNPSSGVHQEYMMPVLSKSPVTTNSEEYMPPPSPNNFPEYNMSQAPTYESFYEPISPHPSSKTAMQENNLITKKEALSKRSPLPKEQEVDALTNLLVQSITDSQDLDVFGICVKCGERISGENAGCTAMGNTYHVHCFTCQRCNVNLQGKPFYDVENEPYCEADYYDTLEKCCVCRKIILDRILRATGKPYHPTCFSCVECGKSLDGIPFTVDAMNQIHCIDDFHKKFAPRCCVCELPIMPEEGQEETVRVVALDRSFHVKCYRCEDCGLLLSSEAEGRGCYPLDDHILCKTCNARRVRLLTNVMTTDL
Summary
Uniprot
H9IXA2
A0A1E1W6Y1
S4NY13
A0A2A4K5Z5
A0A0L7L412
A0A194QEV7
+ More
A0A212FGX6 A0A2H1V5H7 A0A2Z5U7Z0 A0A1Y1MR98 A0A1W4XMI9 T1DKK8 Q171B9 A0A1S4FHA2 A0A182F561 A0A2M4BG54 A0A2M4AQ32 A0A182GEL0 A0A1B0CX89 D2A3T5 W5J4L5 A0A2M4BG61 A0A182J501 T1H9J0 A0A336M6W7 A0A084VPH1 A0A2M3ZAY4 A0A2M3Z2H6 A0A2M3Z3D3 A0A2M3Z2N2 A0A0A9XTN0 A0A023F380 A0A0V0G9Q6 A0A069DTQ5 A0A2M3Z281 A0A2M3ZA16 A0A2M3Z320 A0A224XPZ5 A0A1L8DLK4 A0A1L8DLT3 A0A1B6DVF7 A0A182Q5P5 A0A182NEF9 A0A182VUY1 A0A182MHQ7 A0A182S4R8 U5EQQ5 A0A182Y5V3 Q7PSB2 A0A182T6N0 A0A182K0N6 V5GKM8 A0A3Q0JIW9 A0A182PD82 A0A0C9PZR3 A0A182UNF5 A0A0J7KRI2 A0A2A3EC25 A0A182IE52 A0A088ARC6 A0A182L5M0 A0A293LXZ7 A0A195CCW0 A0A182X7L5 A0A0M9A193 E0VNM2 A0A131Y0K9 A0A1B6LA22 A0A1B6HLK0 A0A154PIT7 A0A195DP55 A0A1A9XW04 N6U3C0 A0A131XHM1 A0A147BVL2 A0A0L7REP7 A0A1B0AP46 E2AP54 F4W4G5 A0A151WZG6 A0A195EZ42 A0A158P364 A0A224Z213 A0A131YYB4 B0W5S3 A0A195BIH7 E2C3I3 A0A026WV63 T1JE70 A0A1A9UXH7 A0A1E1XBJ0 A0A1B0FII6 A0A1A9Z7Q3 A0A1Q3F6A1 A0A1Q3F6F5 A0A1E1XM69
A0A212FGX6 A0A2H1V5H7 A0A2Z5U7Z0 A0A1Y1MR98 A0A1W4XMI9 T1DKK8 Q171B9 A0A1S4FHA2 A0A182F561 A0A2M4BG54 A0A2M4AQ32 A0A182GEL0 A0A1B0CX89 D2A3T5 W5J4L5 A0A2M4BG61 A0A182J501 T1H9J0 A0A336M6W7 A0A084VPH1 A0A2M3ZAY4 A0A2M3Z2H6 A0A2M3Z3D3 A0A2M3Z2N2 A0A0A9XTN0 A0A023F380 A0A0V0G9Q6 A0A069DTQ5 A0A2M3Z281 A0A2M3ZA16 A0A2M3Z320 A0A224XPZ5 A0A1L8DLK4 A0A1L8DLT3 A0A1B6DVF7 A0A182Q5P5 A0A182NEF9 A0A182VUY1 A0A182MHQ7 A0A182S4R8 U5EQQ5 A0A182Y5V3 Q7PSB2 A0A182T6N0 A0A182K0N6 V5GKM8 A0A3Q0JIW9 A0A182PD82 A0A0C9PZR3 A0A182UNF5 A0A0J7KRI2 A0A2A3EC25 A0A182IE52 A0A088ARC6 A0A182L5M0 A0A293LXZ7 A0A195CCW0 A0A182X7L5 A0A0M9A193 E0VNM2 A0A131Y0K9 A0A1B6LA22 A0A1B6HLK0 A0A154PIT7 A0A195DP55 A0A1A9XW04 N6U3C0 A0A131XHM1 A0A147BVL2 A0A0L7REP7 A0A1B0AP46 E2AP54 F4W4G5 A0A151WZG6 A0A195EZ42 A0A158P364 A0A224Z213 A0A131YYB4 B0W5S3 A0A195BIH7 E2C3I3 A0A026WV63 T1JE70 A0A1A9UXH7 A0A1E1XBJ0 A0A1B0FII6 A0A1A9Z7Q3 A0A1Q3F6A1 A0A1Q3F6F5 A0A1E1XM69
Pubmed
19121390
23622113
26227816
26354079
22118469
28004739
+ More
17510324 26483478 18362917 19820115 20920257 23761445 24438588 25401762 26823975 25474469 26334808 25244985 12364791 20966253 20566863 23537049 28049606 29652888 20798317 21719571 21347285 28797301 26830274 24508170 30249741 28503490 29209593
17510324 26483478 18362917 19820115 20920257 23761445 24438588 25401762 26823975 25474469 26334808 25244985 12364791 20966253 20566863 23537049 28049606 29652888 20798317 21719571 21347285 28797301 26830274 24508170 30249741 28503490 29209593
EMBL
BABH01018077
GDQN01008298
GDQN01004232
JAT82756.1
JAT86822.1
GAIX01013980
+ More
JAA78580.1 NWSH01000136 PCG79193.1 JTDY01003160 KOB70039.1 KQ459053 KPJ03954.1 AGBW02008579 OWR53001.1 ODYU01000592 SOQ35642.1 AP017509 BBB06821.1 GEZM01023896 JAV88159.1 GAMD01000901 JAB00690.1 CH477460 EAT40570.1 GGFJ01002898 MBW52039.1 GGFK01009559 MBW42880.1 JXUM01058229 KQ561993 KXJ76959.1 AJWK01033441 AJWK01033442 KQ971348 EFA05566.2 ADMH02002089 ETN59402.1 GGFJ01002899 MBW52040.1 ACPB03022132 UFQT01000212 SSX21768.1 ATLV01015004 KE524999 KFB39865.1 GGFM01004955 MBW25706.1 GGFM01001970 MBW22721.1 GGFM01002271 MBW23022.1 GGFM01002023 MBW22774.1 GBHO01023114 GDHC01008073 JAG20490.1 JAQ10556.1 GBBI01003009 JAC15703.1 GECL01001536 JAP04588.1 GBGD01001431 JAC87458.1 GGFM01001862 MBW22613.1 GGFM01004591 MBW25342.1 GGFM01002087 MBW22838.1 GFTR01006357 JAW10069.1 GFDF01006748 JAV07336.1 GFDF01006749 JAV07335.1 GEDC01031088 GEDC01007657 JAS06210.1 JAS29641.1 AXCN02000859 AXCM01006443 GANO01004152 JAB55719.1 AAAB01008839 EAA05908.5 GALX01003832 JAB64634.1 GBYB01007048 JAG76815.1 LBMM01003957 KMQ92953.1 KZ288308 PBC28749.1 APCN01004564 GFWV01008813 MAA33542.1 KQ977935 KYM98699.1 KQ435791 KOX74326.1 DS235340 EEB14978.1 GEFM01004385 JAP71411.1 GEBQ01019429 JAT20548.1 GECU01032148 GECU01013051 JAS75558.1 JAS94655.1 KQ434931 KZC11796.1 KQ980713 KYN14269.1 APGK01044250 KB741021 ENN75111.1 GEFH01001968 JAP66613.1 GEGO01001029 JAR94375.1 KQ414608 KOC69427.1 JXJN01001129 GL441452 EFN64790.1 GL887532 EGI70860.1 KQ982649 KYQ53091.1 KQ981905 KYN33575.1 ADTU01007761 GFPF01009457 MAA20603.1 GEDV01005391 JAP83166.1 DS231844 EDS35726.1 KQ976467 KYM84172.1 GL452320 EFN77521.1 KK107087 QOIP01000001 EZA59940.1 RLU27270.1 JH432116 GFAC01002620 JAT96568.1 CCAG010022166 GFDL01011968 JAV23077.1 GFDL01011987 JAV23058.1 GFAA01003024 JAU00411.1
JAA78580.1 NWSH01000136 PCG79193.1 JTDY01003160 KOB70039.1 KQ459053 KPJ03954.1 AGBW02008579 OWR53001.1 ODYU01000592 SOQ35642.1 AP017509 BBB06821.1 GEZM01023896 JAV88159.1 GAMD01000901 JAB00690.1 CH477460 EAT40570.1 GGFJ01002898 MBW52039.1 GGFK01009559 MBW42880.1 JXUM01058229 KQ561993 KXJ76959.1 AJWK01033441 AJWK01033442 KQ971348 EFA05566.2 ADMH02002089 ETN59402.1 GGFJ01002899 MBW52040.1 ACPB03022132 UFQT01000212 SSX21768.1 ATLV01015004 KE524999 KFB39865.1 GGFM01004955 MBW25706.1 GGFM01001970 MBW22721.1 GGFM01002271 MBW23022.1 GGFM01002023 MBW22774.1 GBHO01023114 GDHC01008073 JAG20490.1 JAQ10556.1 GBBI01003009 JAC15703.1 GECL01001536 JAP04588.1 GBGD01001431 JAC87458.1 GGFM01001862 MBW22613.1 GGFM01004591 MBW25342.1 GGFM01002087 MBW22838.1 GFTR01006357 JAW10069.1 GFDF01006748 JAV07336.1 GFDF01006749 JAV07335.1 GEDC01031088 GEDC01007657 JAS06210.1 JAS29641.1 AXCN02000859 AXCM01006443 GANO01004152 JAB55719.1 AAAB01008839 EAA05908.5 GALX01003832 JAB64634.1 GBYB01007048 JAG76815.1 LBMM01003957 KMQ92953.1 KZ288308 PBC28749.1 APCN01004564 GFWV01008813 MAA33542.1 KQ977935 KYM98699.1 KQ435791 KOX74326.1 DS235340 EEB14978.1 GEFM01004385 JAP71411.1 GEBQ01019429 JAT20548.1 GECU01032148 GECU01013051 JAS75558.1 JAS94655.1 KQ434931 KZC11796.1 KQ980713 KYN14269.1 APGK01044250 KB741021 ENN75111.1 GEFH01001968 JAP66613.1 GEGO01001029 JAR94375.1 KQ414608 KOC69427.1 JXJN01001129 GL441452 EFN64790.1 GL887532 EGI70860.1 KQ982649 KYQ53091.1 KQ981905 KYN33575.1 ADTU01007761 GFPF01009457 MAA20603.1 GEDV01005391 JAP83166.1 DS231844 EDS35726.1 KQ976467 KYM84172.1 GL452320 EFN77521.1 KK107087 QOIP01000001 EZA59940.1 RLU27270.1 JH432116 GFAC01002620 JAT96568.1 CCAG010022166 GFDL01011968 JAV23077.1 GFDL01011987 JAV23058.1 GFAA01003024 JAU00411.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000007151
UP000192223
+ More
UP000008820 UP000069272 UP000069940 UP000249989 UP000092461 UP000007266 UP000000673 UP000075880 UP000015103 UP000030765 UP000075886 UP000075884 UP000075920 UP000075883 UP000075900 UP000076408 UP000007062 UP000075901 UP000075881 UP000079169 UP000075885 UP000075903 UP000036403 UP000242457 UP000075840 UP000005203 UP000075882 UP000078542 UP000076407 UP000053105 UP000009046 UP000076502 UP000078492 UP000092443 UP000019118 UP000053825 UP000092460 UP000000311 UP000007755 UP000075809 UP000078541 UP000005205 UP000002320 UP000078540 UP000008237 UP000053097 UP000279307 UP000078200 UP000092444 UP000092445
UP000008820 UP000069272 UP000069940 UP000249989 UP000092461 UP000007266 UP000000673 UP000075880 UP000015103 UP000030765 UP000075886 UP000075884 UP000075920 UP000075883 UP000075900 UP000076408 UP000007062 UP000075901 UP000075881 UP000079169 UP000075885 UP000075903 UP000036403 UP000242457 UP000075840 UP000005203 UP000075882 UP000078542 UP000076407 UP000053105 UP000009046 UP000076502 UP000078492 UP000092443 UP000019118 UP000053825 UP000092460 UP000000311 UP000007755 UP000075809 UP000078541 UP000005205 UP000002320 UP000078540 UP000008237 UP000053097 UP000279307 UP000078200 UP000092444 UP000092445
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IXA2
A0A1E1W6Y1
S4NY13
A0A2A4K5Z5
A0A0L7L412
A0A194QEV7
+ More
A0A212FGX6 A0A2H1V5H7 A0A2Z5U7Z0 A0A1Y1MR98 A0A1W4XMI9 T1DKK8 Q171B9 A0A1S4FHA2 A0A182F561 A0A2M4BG54 A0A2M4AQ32 A0A182GEL0 A0A1B0CX89 D2A3T5 W5J4L5 A0A2M4BG61 A0A182J501 T1H9J0 A0A336M6W7 A0A084VPH1 A0A2M3ZAY4 A0A2M3Z2H6 A0A2M3Z3D3 A0A2M3Z2N2 A0A0A9XTN0 A0A023F380 A0A0V0G9Q6 A0A069DTQ5 A0A2M3Z281 A0A2M3ZA16 A0A2M3Z320 A0A224XPZ5 A0A1L8DLK4 A0A1L8DLT3 A0A1B6DVF7 A0A182Q5P5 A0A182NEF9 A0A182VUY1 A0A182MHQ7 A0A182S4R8 U5EQQ5 A0A182Y5V3 Q7PSB2 A0A182T6N0 A0A182K0N6 V5GKM8 A0A3Q0JIW9 A0A182PD82 A0A0C9PZR3 A0A182UNF5 A0A0J7KRI2 A0A2A3EC25 A0A182IE52 A0A088ARC6 A0A182L5M0 A0A293LXZ7 A0A195CCW0 A0A182X7L5 A0A0M9A193 E0VNM2 A0A131Y0K9 A0A1B6LA22 A0A1B6HLK0 A0A154PIT7 A0A195DP55 A0A1A9XW04 N6U3C0 A0A131XHM1 A0A147BVL2 A0A0L7REP7 A0A1B0AP46 E2AP54 F4W4G5 A0A151WZG6 A0A195EZ42 A0A158P364 A0A224Z213 A0A131YYB4 B0W5S3 A0A195BIH7 E2C3I3 A0A026WV63 T1JE70 A0A1A9UXH7 A0A1E1XBJ0 A0A1B0FII6 A0A1A9Z7Q3 A0A1Q3F6A1 A0A1Q3F6F5 A0A1E1XM69
A0A212FGX6 A0A2H1V5H7 A0A2Z5U7Z0 A0A1Y1MR98 A0A1W4XMI9 T1DKK8 Q171B9 A0A1S4FHA2 A0A182F561 A0A2M4BG54 A0A2M4AQ32 A0A182GEL0 A0A1B0CX89 D2A3T5 W5J4L5 A0A2M4BG61 A0A182J501 T1H9J0 A0A336M6W7 A0A084VPH1 A0A2M3ZAY4 A0A2M3Z2H6 A0A2M3Z3D3 A0A2M3Z2N2 A0A0A9XTN0 A0A023F380 A0A0V0G9Q6 A0A069DTQ5 A0A2M3Z281 A0A2M3ZA16 A0A2M3Z320 A0A224XPZ5 A0A1L8DLK4 A0A1L8DLT3 A0A1B6DVF7 A0A182Q5P5 A0A182NEF9 A0A182VUY1 A0A182MHQ7 A0A182S4R8 U5EQQ5 A0A182Y5V3 Q7PSB2 A0A182T6N0 A0A182K0N6 V5GKM8 A0A3Q0JIW9 A0A182PD82 A0A0C9PZR3 A0A182UNF5 A0A0J7KRI2 A0A2A3EC25 A0A182IE52 A0A088ARC6 A0A182L5M0 A0A293LXZ7 A0A195CCW0 A0A182X7L5 A0A0M9A193 E0VNM2 A0A131Y0K9 A0A1B6LA22 A0A1B6HLK0 A0A154PIT7 A0A195DP55 A0A1A9XW04 N6U3C0 A0A131XHM1 A0A147BVL2 A0A0L7REP7 A0A1B0AP46 E2AP54 F4W4G5 A0A151WZG6 A0A195EZ42 A0A158P364 A0A224Z213 A0A131YYB4 B0W5S3 A0A195BIH7 E2C3I3 A0A026WV63 T1JE70 A0A1A9UXH7 A0A1E1XBJ0 A0A1B0FII6 A0A1A9Z7Q3 A0A1Q3F6A1 A0A1Q3F6F5 A0A1E1XM69
PDB
2DLO
E-value=1.8815e-20,
Score=244
Ontologies
GO
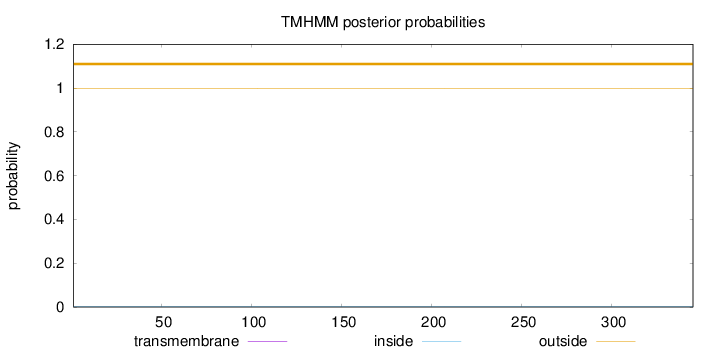
Topology
Length:
345
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00106
outside
1 - 345
Population Genetic Test Statistics
Pi
218.946427
Theta
185.829057
Tajima's D
0.630716
CLR
0
CSRT
0.552222388880556
Interpretation
Uncertain
