Gene
KWMTBOMO11424
Pre Gene Modal
BGIBMGA001882
Annotation
PREDICTED:_potassium/sodium_hyperpolarization-activated_cyclic_nucleotide-gated_channel_2_isoform_X12_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.756
Sequence
CDS
ATGGGAGGCTACCTAAAATTCTTGCCGGAACACGACAGATTTTACTGGGACTTATGCATGCTGCTACTTCTTGTGGCGAATTTAATCATCCTACCAGTTGCCATCTCCTTCTTCAACGATGACCTTAGCACGCGCTGGATTGCCTTCAATTGTCTGTCCGATACCATATTTCTAATAGATATCGTTGTTAATTTTAGAACAGGAATAATGCAACAAGATAATGCAGAGCAAGTAATACTAGATCCAAAGTTAATAGCGAAACACTATTTGAGGACTTGGTTCTTCCTGGACCTCATCTCTAGTATACCACTAGATTATATATTTTTGATCTTCAATCAGGTAAACGATTTCAGTGAAAACTTTCAAATACTACATGCGGGTCGCGCGCTGAGGATACTCCGATTAGCAAAGCTGCTATCATTGGTTCGACTGCTCCGTCTCTCCAGGCTCGTCCGATATGTCTCACAATGGGAGGAAGTATATTTTCTCAACATGGCTTCCGTGTTCATGCGGATATTCAACTTAATATGTATGATGCTGTTGATCGGCCATTGGTCCGGATGTCTACAGTTCCTGGTTCCTATGCTGCAAGGATTTCCACCAAACTCGTGGGTTGCTATAAACGAGTTACAGGAGGCTTTTTGGTTAGAACAATATTCATGGGCGCTGTTCAAGGCAATGTCACATATGTTGTGCATCGGTTACGGACGGTTCCCTCCTCAATCACTCACCGACATGTGGCTCACGATGCTCTCGATGATATCAGGAGCCACTTGCTACGCGCTCTTCCTAGGTCACGCAACCAATCTCATCCAGAGTCTCGACTCTTCACGCAGGCAATACCGTGAAAAGGTGAAACAAGTAGAAGAGTACATGGCGTATCGCAAATTACCACGTGAAATGCGTCAACGTATCACCGAGTACTTCGAACACAGATACCAAGGGAAATTCTTCGATGAGGAAGTAATTTTGGGCGAGCTGAGCGAAAAACTGCGCGAAGATGTCATCAACTACAACTGTCGCTCACTTGTGGCCTCTGTACCTTTCTTTGCAAACGCCGACTCCAACTTTGTGTCAGATGTCGTCACCAAACTACGCTATGAAGTGTTTCAGCCTGGTGACATTATCATAAAGGAAGGAACAATAGGAAATAAAATGTATTTCATCCAAGAAGGCATCGTAGATATAGTCATGGCCAACGGTGAAGTTGCCACCAGTCTATCCGACGGTTCGTACTTCGGAGAAATCTGCTTGCTAACAAACGCTCGCAGAGTAGCATCCGTCCGCGCCGAGACGTACTGCAATCTGTTCTCTTTGTCGGTGGATCATTTCAACGCCGTGCTAGACCAGTATCCGTTGATGCGACGCACGATGGAAAGTGTCGCAGCCGAACGACTGAACAAGATCGGCAAAAACCCGAATCTGGTGGCGCACCGCGAAGACGACACGACATCCGAAGGGAACACGATCAACGCAGTGGTTAACGCTCTCGCGGCAGAAGCAGAGCACGTGAGTCTGTCAGACGACAGTGTGGCACGACTGTCGGAGAGGTCGCTGGGGCTAGCACTGCAACCGCTGCAGGCGGCATCGTGTCGCATGGCAGGTGTGGCCCTACCGGGGCTGGGCGTAGCGGCTGCGCTGCCGCGTCCCAAGTCGGAGCACGACTTCAGTTCGACGCAGGCTCAGCCGCCGTCGTTGTCGTCGGTCGGAGCCGCCTTCCACAAATCGGACGCGGGCATCGCACCCTGA
Protein
MGGYLKFLPEHDRFYWDLCMLLLLVANLIILPVAISFFNDDLSTRWIAFNCLSDTIFLIDIVVNFRTGIMQQDNAEQVILDPKLIAKHYLRTWFFLDLISSIPLDYIFLIFNQVNDFSENFQILHAGRALRILRLAKLLSLVRLLRLSRLVRYVSQWEEVYFLNMASVFMRIFNLICMMLLIGHWSGCLQFLVPMLQGFPPNSWVAINELQEAFWLEQYSWALFKAMSHMLCIGYGRFPPQSLTDMWLTMLSMISGATCYALFLGHATNLIQSLDSSRRQYREKVKQVEEYMAYRKLPREMRQRITEYFEHRYQGKFFDEEVILGELSEKLREDVINYNCRSLVASVPFFANADSNFVSDVVTKLRYEVFQPGDIIIKEGTIGNKMYFIQEGIVDIVMANGEVATSLSDGSYFGEICLLTNARRVASVRAETYCNLFSLSVDHFNAVLDQYPLMRRTMESVAAERLNKIGKNPNLVAHREDDTTSEGNTINAVVNALAAEAEHVSLSDDSVARLSERSLGLALQPLQAASCRMAGVALPGLGVAAALPRPKSEHDFSSTQAQPPSLSSVGAAFHKSDAGIAP
Summary
Uniprot
A0A2A4K4K4
A0A2A4K4L7
O96777
S4PAD1
K7INL3
A0A158NZ19
+ More
A0A2A3EAG6 Q6WL04 A0A1Y1L055 A0A1Y1KXP8 K7INL2 A0A139WF05 A0A154P706 Q5XQT6 A0A151WKF2 A0A088A929 A0A232EZ95 A0A139WF98 A0A026W9G0 F4W9U9 A0A084WS95 A0A034V7Z9 A0A182F6Z2 A0A0K8VQZ6 B7YZE7 A1Z9N8 A0A1J1HRM1 A0A0Q5VWH7 A0A1W4VZ70 A0A0R3NQ18 A0A0R3NXD6 A0A0P9C033 A0A0Q9X0V2 A0A0Q5VMJ9 B4QFF5 A0A0R3NW28 Q56JI0 A0A0R1DQM2 A0A0N8P0E5 Q16ZU7 A0A1I8MGZ7 A0A0R1DQJ9 A0A0Q9WXF4 A0A1W4WBE4 A0A1I8MH01 A0A0R3NVR5 A0A3B0J0U5 A0A3B0J4W2 N6VFV5 A0A0R3NQ48 A0A1I8PZB0 A0A034V5W6 A0A034V5W1 A0A1I8PZ82 Q9Y1J9 W5J915 A0A0A1XB94 W8BX37 E0W2H0 F7VJU3 A0A1J1HRQ0 A1Z9P0 A0A0Q9WYQ8 Q56JH8 A0A0Q5VNV9 A0A0P8ZR17 Q56JH9 A0A0R1DXL0 A0A1J1HW14 A0A034V8L7 B4HRA8 F5HIW0 B4KSK8 A1Z9N7 A0A0Q9WNN2 A0A0Q5VNM9 A0A3B0J1B4 A0A0R3NPR9 Q28X53 A0A0R3NPY8 Q56JH7 B7YZE8 A0A0R3NWD5 A0A0R3NUU7 B4P738 A0A0P8XQQ5 B4MJM3 A0A1W4WA06 A0A0J9RCI0 A0A1I8MGZ4 A0A1B6HCW2 A0A1W4WAY6 A0A0R1DQU9 B3MDZ1 B3NR62 A0A1I8MGY4 A0A1I8MGZ2 A0A0R3NPS5 A0A1I8MGZ6 A0A1I8PZ81
A0A2A3EAG6 Q6WL04 A0A1Y1L055 A0A1Y1KXP8 K7INL2 A0A139WF05 A0A154P706 Q5XQT6 A0A151WKF2 A0A088A929 A0A232EZ95 A0A139WF98 A0A026W9G0 F4W9U9 A0A084WS95 A0A034V7Z9 A0A182F6Z2 A0A0K8VQZ6 B7YZE7 A1Z9N8 A0A1J1HRM1 A0A0Q5VWH7 A0A1W4VZ70 A0A0R3NQ18 A0A0R3NXD6 A0A0P9C033 A0A0Q9X0V2 A0A0Q5VMJ9 B4QFF5 A0A0R3NW28 Q56JI0 A0A0R1DQM2 A0A0N8P0E5 Q16ZU7 A0A1I8MGZ7 A0A0R1DQJ9 A0A0Q9WXF4 A0A1W4WBE4 A0A1I8MH01 A0A0R3NVR5 A0A3B0J0U5 A0A3B0J4W2 N6VFV5 A0A0R3NQ48 A0A1I8PZB0 A0A034V5W6 A0A034V5W1 A0A1I8PZ82 Q9Y1J9 W5J915 A0A0A1XB94 W8BX37 E0W2H0 F7VJU3 A0A1J1HRQ0 A1Z9P0 A0A0Q9WYQ8 Q56JH8 A0A0Q5VNV9 A0A0P8ZR17 Q56JH9 A0A0R1DXL0 A0A1J1HW14 A0A034V8L7 B4HRA8 F5HIW0 B4KSK8 A1Z9N7 A0A0Q9WNN2 A0A0Q5VNM9 A0A3B0J1B4 A0A0R3NPR9 Q28X53 A0A0R3NPY8 Q56JH7 B7YZE8 A0A0R3NWD5 A0A0R3NUU7 B4P738 A0A0P8XQQ5 B4MJM3 A0A1W4WA06 A0A0J9RCI0 A0A1I8MGZ4 A0A1B6HCW2 A0A1W4WAY6 A0A0R1DQU9 B3MDZ1 B3NR62 A0A1I8MGY4 A0A1I8MGZ2 A0A0R3NPS5 A0A1I8MGZ6 A0A1I8PZ81
Pubmed
10319439
23622113
20075255
21347285
14563363
28004739
+ More
18362917 19820115 15388341 28648823 24508170 21719571 24438588 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 15632085 15804582 17550304 17510324 25315136 12491074 20920257 23761445 25830018 24495485 20566863 18057021 26109357 26109356 12364791 22936249
18362917 19820115 15388341 28648823 24508170 21719571 24438588 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 15632085 15804582 17550304 17510324 25315136 12491074 20920257 23761445 25830018 24495485 20566863 18057021 26109357 26109356 12364791 22936249
EMBL
NWSH01000136
PCG79185.1
PCG79187.1
AJ012664
CAA10110.1
GAIX01005081
+ More
JAA87479.1 ADTU01000847 ADTU01000848 ADTU01000849 ADTU01000850 ADTU01000851 ADTU01000852 ADTU01000853 ADTU01000854 ADTU01000855 ADTU01000856 KZ288318 PBC28282.1 AY280848 AAQ16312.1 GEZM01070976 JAV66151.1 GEZM01070975 JAV66152.1 KQ971354 KYB26550.1 KQ434829 KZC07726.1 AY739658 AAU85297.1 KQ983012 KYQ48353.1 NNAY01001523 OXU23701.1 KYB26549.1 KK107321 EZA52675.1 GL888033 EGI69085.1 ATLV01026403 KE525413 KFB53089.1 GAKP01021057 JAC37895.1 GDHF01011008 JAI41306.1 AE013599 ACL83121.1 ABC66058.1 CVRI01000020 CRK90703.1 CH954179 KQS62633.1 CM000071 KRT03122.1 KRT03126.1 CH902619 KPU76898.1 CH963846 KRF97533.1 KQS62631.1 CM000362 EDX07065.1 KRT03120.1 AY928069 AAX78393.1 CM000158 KRJ99559.1 KPU76900.1 CH477484 EAT40174.1 KRJ99557.1 KRF97534.1 KRT03125.1 OUUW01000001 SPP74654.1 SPP74653.1 ENO01935.1 KRT03124.1 GAKP01021053 JAC37899.1 GAKP01021058 JAC37894.1 AF124300 AAD42059.1 ADMH02001843 ETN60932.1 GBXI01005648 JAD08644.1 GAMC01008724 JAB97831.1 DS235878 EEB19826.1 BT128743 AEH59646.1 CRK90701.1 ABC66060.2 KRF97532.1 AY928071 AAX78395.1 KQS62632.1 KQS62634.1 KQS62636.1 KPU76895.1 KPU76899.1 AY928070 AAX78394.1 ABC66059.1 AFH08072.1 AFH08073.1 AFH08074.1 KRJ99558.1 KRJ99561.1 KRJ99562.1 CRK90702.1 GAKP01021054 JAC37898.1 CH480816 EDW47837.1 AAAB01008799 EGK96221.1 CH933808 EDW09513.2 ABC66057.1 KRF97530.1 KQS62628.1 SPP74655.1 KRT03121.1 EAL26463.3 KRT03127.1 AY928072 AAX78396.1 ACL83122.1 KRT03123.1 KRT03119.1 EDW91003.2 KPU76893.1 EDW72312.2 CM002911 KMY93738.1 GECU01035169 JAS72537.1 KRJ99560.1 EDV37536.2 EDV56051.1 KRT03118.1
JAA87479.1 ADTU01000847 ADTU01000848 ADTU01000849 ADTU01000850 ADTU01000851 ADTU01000852 ADTU01000853 ADTU01000854 ADTU01000855 ADTU01000856 KZ288318 PBC28282.1 AY280848 AAQ16312.1 GEZM01070976 JAV66151.1 GEZM01070975 JAV66152.1 KQ971354 KYB26550.1 KQ434829 KZC07726.1 AY739658 AAU85297.1 KQ983012 KYQ48353.1 NNAY01001523 OXU23701.1 KYB26549.1 KK107321 EZA52675.1 GL888033 EGI69085.1 ATLV01026403 KE525413 KFB53089.1 GAKP01021057 JAC37895.1 GDHF01011008 JAI41306.1 AE013599 ACL83121.1 ABC66058.1 CVRI01000020 CRK90703.1 CH954179 KQS62633.1 CM000071 KRT03122.1 KRT03126.1 CH902619 KPU76898.1 CH963846 KRF97533.1 KQS62631.1 CM000362 EDX07065.1 KRT03120.1 AY928069 AAX78393.1 CM000158 KRJ99559.1 KPU76900.1 CH477484 EAT40174.1 KRJ99557.1 KRF97534.1 KRT03125.1 OUUW01000001 SPP74654.1 SPP74653.1 ENO01935.1 KRT03124.1 GAKP01021053 JAC37899.1 GAKP01021058 JAC37894.1 AF124300 AAD42059.1 ADMH02001843 ETN60932.1 GBXI01005648 JAD08644.1 GAMC01008724 JAB97831.1 DS235878 EEB19826.1 BT128743 AEH59646.1 CRK90701.1 ABC66060.2 KRF97532.1 AY928071 AAX78395.1 KQS62632.1 KQS62634.1 KQS62636.1 KPU76895.1 KPU76899.1 AY928070 AAX78394.1 ABC66059.1 AFH08072.1 AFH08073.1 AFH08074.1 KRJ99558.1 KRJ99561.1 KRJ99562.1 CRK90702.1 GAKP01021054 JAC37898.1 CH480816 EDW47837.1 AAAB01008799 EGK96221.1 CH933808 EDW09513.2 ABC66057.1 KRF97530.1 KQS62628.1 SPP74655.1 KRT03121.1 EAL26463.3 KRT03127.1 AY928072 AAX78396.1 ACL83122.1 KRT03123.1 KRT03119.1 EDW91003.2 KPU76893.1 EDW72312.2 CM002911 KMY93738.1 GECU01035169 JAS72537.1 KRJ99560.1 EDV37536.2 EDV56051.1 KRT03118.1
Proteomes
UP000218220
UP000002358
UP000005205
UP000242457
UP000007266
UP000076502
+ More
UP000075809 UP000005203 UP000215335 UP000053097 UP000007755 UP000030765 UP000069272 UP000000803 UP000183832 UP000008711 UP000192221 UP000001819 UP000007801 UP000007798 UP000000304 UP000002282 UP000008820 UP000095301 UP000268350 UP000095300 UP000000673 UP000009046 UP000001292 UP000007062 UP000009192
UP000075809 UP000005203 UP000215335 UP000053097 UP000007755 UP000030765 UP000069272 UP000000803 UP000183832 UP000008711 UP000192221 UP000001819 UP000007801 UP000007798 UP000000304 UP000002282 UP000008820 UP000095301 UP000268350 UP000095300 UP000000673 UP000009046 UP000001292 UP000007062 UP000009192
Interpro
SUPFAM
SSF51206
SSF51206
Gene 3D
CDD
ProteinModelPortal
A0A2A4K4K4
A0A2A4K4L7
O96777
S4PAD1
K7INL3
A0A158NZ19
+ More
A0A2A3EAG6 Q6WL04 A0A1Y1L055 A0A1Y1KXP8 K7INL2 A0A139WF05 A0A154P706 Q5XQT6 A0A151WKF2 A0A088A929 A0A232EZ95 A0A139WF98 A0A026W9G0 F4W9U9 A0A084WS95 A0A034V7Z9 A0A182F6Z2 A0A0K8VQZ6 B7YZE7 A1Z9N8 A0A1J1HRM1 A0A0Q5VWH7 A0A1W4VZ70 A0A0R3NQ18 A0A0R3NXD6 A0A0P9C033 A0A0Q9X0V2 A0A0Q5VMJ9 B4QFF5 A0A0R3NW28 Q56JI0 A0A0R1DQM2 A0A0N8P0E5 Q16ZU7 A0A1I8MGZ7 A0A0R1DQJ9 A0A0Q9WXF4 A0A1W4WBE4 A0A1I8MH01 A0A0R3NVR5 A0A3B0J0U5 A0A3B0J4W2 N6VFV5 A0A0R3NQ48 A0A1I8PZB0 A0A034V5W6 A0A034V5W1 A0A1I8PZ82 Q9Y1J9 W5J915 A0A0A1XB94 W8BX37 E0W2H0 F7VJU3 A0A1J1HRQ0 A1Z9P0 A0A0Q9WYQ8 Q56JH8 A0A0Q5VNV9 A0A0P8ZR17 Q56JH9 A0A0R1DXL0 A0A1J1HW14 A0A034V8L7 B4HRA8 F5HIW0 B4KSK8 A1Z9N7 A0A0Q9WNN2 A0A0Q5VNM9 A0A3B0J1B4 A0A0R3NPR9 Q28X53 A0A0R3NPY8 Q56JH7 B7YZE8 A0A0R3NWD5 A0A0R3NUU7 B4P738 A0A0P8XQQ5 B4MJM3 A0A1W4WA06 A0A0J9RCI0 A0A1I8MGZ4 A0A1B6HCW2 A0A1W4WAY6 A0A0R1DQU9 B3MDZ1 B3NR62 A0A1I8MGY4 A0A1I8MGZ2 A0A0R3NPS5 A0A1I8MGZ6 A0A1I8PZ81
A0A2A3EAG6 Q6WL04 A0A1Y1L055 A0A1Y1KXP8 K7INL2 A0A139WF05 A0A154P706 Q5XQT6 A0A151WKF2 A0A088A929 A0A232EZ95 A0A139WF98 A0A026W9G0 F4W9U9 A0A084WS95 A0A034V7Z9 A0A182F6Z2 A0A0K8VQZ6 B7YZE7 A1Z9N8 A0A1J1HRM1 A0A0Q5VWH7 A0A1W4VZ70 A0A0R3NQ18 A0A0R3NXD6 A0A0P9C033 A0A0Q9X0V2 A0A0Q5VMJ9 B4QFF5 A0A0R3NW28 Q56JI0 A0A0R1DQM2 A0A0N8P0E5 Q16ZU7 A0A1I8MGZ7 A0A0R1DQJ9 A0A0Q9WXF4 A0A1W4WBE4 A0A1I8MH01 A0A0R3NVR5 A0A3B0J0U5 A0A3B0J4W2 N6VFV5 A0A0R3NQ48 A0A1I8PZB0 A0A034V5W6 A0A034V5W1 A0A1I8PZ82 Q9Y1J9 W5J915 A0A0A1XB94 W8BX37 E0W2H0 F7VJU3 A0A1J1HRQ0 A1Z9P0 A0A0Q9WYQ8 Q56JH8 A0A0Q5VNV9 A0A0P8ZR17 Q56JH9 A0A0R1DXL0 A0A1J1HW14 A0A034V8L7 B4HRA8 F5HIW0 B4KSK8 A1Z9N7 A0A0Q9WNN2 A0A0Q5VNM9 A0A3B0J1B4 A0A0R3NPR9 Q28X53 A0A0R3NPY8 Q56JH7 B7YZE8 A0A0R3NWD5 A0A0R3NUU7 B4P738 A0A0P8XQQ5 B4MJM3 A0A1W4WA06 A0A0J9RCI0 A0A1I8MGZ4 A0A1B6HCW2 A0A1W4WAY6 A0A0R1DQU9 B3MDZ1 B3NR62 A0A1I8MGY4 A0A1I8MGZ2 A0A0R3NPS5 A0A1I8MGZ6 A0A1I8PZ81
PDB
6GYO
E-value=5.51558e-160,
Score=1449
Ontologies
GO
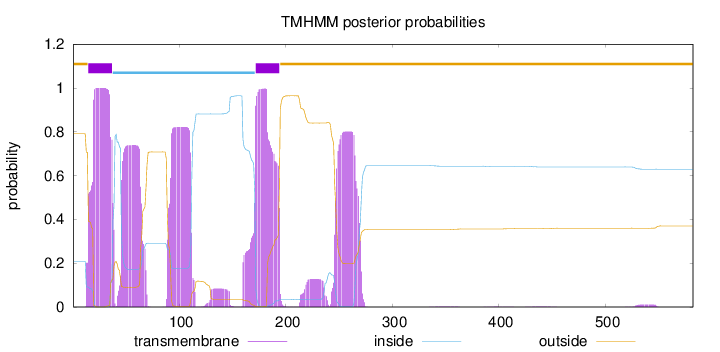
Topology
Length:
582
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
99.6878700000001
Exp number, first 60 AAs:
32.53492
Total prob of N-in:
0.20918
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 171
TMhelix
172 - 194
outside
195 - 582
Population Genetic Test Statistics
Pi
223.711563
Theta
203.856119
Tajima's D
0.180881
CLR
0.055047
CSRT
0.423978801059947
Interpretation
Uncertain
