Gene
KWMTBOMO11422
Pre Gene Modal
BGIBMGA001881
Annotation
Potassium/sodium_hyperpolarization-activated_cyclic_nucleotide-gated_channel_1_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 1.996
Sequence
CDS
ATGTCGCTGAAGCAGGGCAGCGGCACCGGCAAAGTACACTTCGGTGGCCTGGACGATGTGTCCTTGTACGGAACGCCAGTGGAACCAGCACCGCCGGCGCCGGACGCAAAGCAGGGCTTCCTGCGGAACCAGTTGCAAGCCCTCTTCCAGCCCACCGACAACAAACTCGCGATGAAACTATTCGGCTCAAAGAAGGCACTTATGAAGGAGCGCATCAGACAGAAGGCGGCGGGACATTGGGTCATCCATCCGTGCAGTAGTTTCAGGTATGTCCAGATTCAGCACTTTTCTATTTTGTTAATGAGATGA
Protein
MSLKQGSGTGKVHFGGLDDVSLYGTPVEPAPPAPDAKQGFLRNQLQALFQPTDNKLAMKLFGSKKALMKERIRQKAAGHWVIHPCSSFRYVQIQHFSILLMR
Summary
Uniprot
A0A194QEW2
A0A194RFP5
H9IX98
A0A212F2B3
A0A2H1V032
A0A0L7KPU8
+ More
O96777 A0A2A4K4L7 A0A2A4K4K4 F4W9U8 E2BZX9 E2A5U2 V9I8E8 A0A0C9RQN1 A0A067RJN2 V9IAA4 A0A026WCB0 A0A0J7P2B4 A0A1W4XCU8 V9I9T3 A0A2A3EAG6 Q6WL04 A0A1W4XCL5 A0A195CG79 K7INL3 A0A195BRD4 A0A1W4X249 A0A151JPH3 A0A151WKF2 A0A1B6K7N7 A0A158NZ19 A0A0T6BED1 Q5XQT5 N6SRH5 A0A195EY94 A0A1Y1KXL2 A0A1Y1KXP8 A0A1Y1L055 A0A0C9RTG4 A0A0L7R2N3 Q5XQT6 A0A088A929 A0A154P706 K7INL2 A0A232EZ95 A0A1B6KBD0 A0A0P4VVJ6 A0A3L8DY95 E0W2H0 E9IFR6 A0A1Y1L2G8 A0A146L7B4 A0A146LBW4 A0A139WFA2 A0A0V0G8Y1 A0A069DW42 T1HZI8 A0A1B6EXC7 A0A1B6GS55 A0A1B6HGY3 Q06IP4 Q06IQ0 Q06IP2 A0A3Q8TMV0 Q06IP3 Q06IP7 Q06IP5 Q06IP1 Q06IP9 Q6WL05 A0A1B6C0Y8 A0A2P6L6H5 A0A3R7SWG4 A0A0P5M0J4 A0A0P6GXS3 A0A0P5Z2H7 A0A0P5AGG2 A0A0P5ACY5 A0A0P5AA68 A0A0P5EAB5 A0A0P6FJJ4 A0A0N8BGC6 A0A0P4XWK1 A0A0P5ASL0 A0A0P5CIS0 A0A0P5ME34 A0A0P5JMZ8 A0A0P5IDY4 A0A0P6EU10 A0A0P5QHU2 A0A0P5MF30 A0A0P5LT45 A0A0P5MQ71 A0A0P6GA18 A0A0P5H259 A0A0P5ASP2 A0A0P4ZTE3 A0A0P4YXL4 A0A0P5GWF7 A0A0P5DZP8
O96777 A0A2A4K4L7 A0A2A4K4K4 F4W9U8 E2BZX9 E2A5U2 V9I8E8 A0A0C9RQN1 A0A067RJN2 V9IAA4 A0A026WCB0 A0A0J7P2B4 A0A1W4XCU8 V9I9T3 A0A2A3EAG6 Q6WL04 A0A1W4XCL5 A0A195CG79 K7INL3 A0A195BRD4 A0A1W4X249 A0A151JPH3 A0A151WKF2 A0A1B6K7N7 A0A158NZ19 A0A0T6BED1 Q5XQT5 N6SRH5 A0A195EY94 A0A1Y1KXL2 A0A1Y1KXP8 A0A1Y1L055 A0A0C9RTG4 A0A0L7R2N3 Q5XQT6 A0A088A929 A0A154P706 K7INL2 A0A232EZ95 A0A1B6KBD0 A0A0P4VVJ6 A0A3L8DY95 E0W2H0 E9IFR6 A0A1Y1L2G8 A0A146L7B4 A0A146LBW4 A0A139WFA2 A0A0V0G8Y1 A0A069DW42 T1HZI8 A0A1B6EXC7 A0A1B6GS55 A0A1B6HGY3 Q06IP4 Q06IQ0 Q06IP2 A0A3Q8TMV0 Q06IP3 Q06IP7 Q06IP5 Q06IP1 Q06IP9 Q6WL05 A0A1B6C0Y8 A0A2P6L6H5 A0A3R7SWG4 A0A0P5M0J4 A0A0P6GXS3 A0A0P5Z2H7 A0A0P5AGG2 A0A0P5ACY5 A0A0P5AA68 A0A0P5EAB5 A0A0P6FJJ4 A0A0N8BGC6 A0A0P4XWK1 A0A0P5ASL0 A0A0P5CIS0 A0A0P5ME34 A0A0P5JMZ8 A0A0P5IDY4 A0A0P6EU10 A0A0P5QHU2 A0A0P5MF30 A0A0P5LT45 A0A0P5MQ71 A0A0P6GA18 A0A0P5H259 A0A0P5ASP2 A0A0P4ZTE3 A0A0P4YXL4 A0A0P5GWF7 A0A0P5DZP8
Pubmed
EMBL
KQ459053
KPJ03959.1
KQ460436
KPJ14756.1
BABH01018082
AGBW02010766
+ More
OWR47880.1 ODYU01000064 SOQ34197.1 JTDY01007534 KOB65130.1 AJ012664 CAA10110.1 NWSH01000136 PCG79187.1 PCG79185.1 GL888033 EGI69084.1 GL451708 EFN78756.1 GL437059 EFN71214.1 JR036802 AEY57408.1 GBYB01010800 JAG80567.1 KK852430 KDR24022.1 JR036800 AEY57406.1 KK107321 EZA52674.1 LBMM01000321 KMR01382.1 JR036801 AEY57407.1 KZ288318 PBC28282.1 AY280848 AAQ16312.1 KQ977876 KYM99103.1 KQ976423 KYM88774.1 KQ978765 KYN28538.1 KQ983012 KYQ48353.1 GECU01000292 JAT07415.1 ADTU01000847 ADTU01000848 ADTU01000849 ADTU01000850 ADTU01000851 ADTU01000852 ADTU01000853 ADTU01000854 ADTU01000855 ADTU01000856 LJIG01001439 KRT85501.1 AY739659 AAU85298.1 APGK01059354 KB741293 KB631982 ENN70214.1 ERL87627.1 KQ981909 KYN33188.1 GEZM01070974 JAV66153.1 GEZM01070975 JAV66152.1 GEZM01070976 JAV66151.1 GBYB01010781 JAG80548.1 KQ414666 KOC65093.1 AY739658 AAU85297.1 KQ434829 KZC07726.1 NNAY01001523 OXU23701.1 GEBQ01031210 JAT08767.1 GDKW01002950 JAI53645.1 QOIP01000002 RLU25421.1 DS235878 EEB19826.1 GL762889 EFZ20589.1 GEZM01070973 JAV66155.1 GDHC01014301 JAQ04328.1 GDHC01013584 JAQ05045.1 KQ971354 KYB26551.1 GECL01002210 JAP03914.1 GBGD01000828 JAC88061.1 ACPB03006006 ACPB03006007 ACPB03006008 GECZ01027105 JAS42664.1 GECZ01004576 JAS65193.1 GECU01033827 GECU01003848 JAS73879.1 JAT03859.1 DQ865256 ABI94039.1 DQ865250 ABI94033.1 DQ865258 ABI94041.1 MH368784 AZL49143.1 DQ865257 ABI94040.1 DQ865253 ABI94036.1 DQ865255 ABI94038.1 DQ865259 ABI94042.1 DQ865251 ABI94034.1 AY280847 AAQ16311.1 GEDC01030116 JAS07182.1 MWRG01001444 PRD34188.1 QCYY01001360 ROT78575.1 GDIQ01162353 JAK89372.1 GDIQ01039651 JAN55086.1 GDIP01050945 JAM52770.1 GDIP01199469 JAJ23933.1 GDIP01200779 JAJ22623.1 GDIP01201978 JAJ21424.1 GDIP01162922 JAJ60480.1 GDIQ01056438 JAN38299.1 GDIQ01180451 JAK71274.1 GDIP01236770 JAI86631.1 GDIP01209011 JAJ14391.1 GDIP01169965 JAJ53437.1 GDIQ01161010 JAK90715.1 GDIQ01196348 JAK55377.1 GDIQ01216041 JAK35684.1 GDIQ01059086 JAN35651.1 GDIQ01113921 JAL37805.1 GDIQ01180450 JAK71275.1 GDIQ01175789 JAK75936.1 GDIQ01175788 JAK75937.1 GDIQ01036395 JAN58342.1 GDIQ01239046 JAK12679.1 GDIP01194979 JAJ28423.1 GDIP01221774 JAJ01628.1 GDIP01220716 JAJ02686.1 GDIQ01235478 JAK16247.1 GDIP01153421 JAJ69981.1
OWR47880.1 ODYU01000064 SOQ34197.1 JTDY01007534 KOB65130.1 AJ012664 CAA10110.1 NWSH01000136 PCG79187.1 PCG79185.1 GL888033 EGI69084.1 GL451708 EFN78756.1 GL437059 EFN71214.1 JR036802 AEY57408.1 GBYB01010800 JAG80567.1 KK852430 KDR24022.1 JR036800 AEY57406.1 KK107321 EZA52674.1 LBMM01000321 KMR01382.1 JR036801 AEY57407.1 KZ288318 PBC28282.1 AY280848 AAQ16312.1 KQ977876 KYM99103.1 KQ976423 KYM88774.1 KQ978765 KYN28538.1 KQ983012 KYQ48353.1 GECU01000292 JAT07415.1 ADTU01000847 ADTU01000848 ADTU01000849 ADTU01000850 ADTU01000851 ADTU01000852 ADTU01000853 ADTU01000854 ADTU01000855 ADTU01000856 LJIG01001439 KRT85501.1 AY739659 AAU85298.1 APGK01059354 KB741293 KB631982 ENN70214.1 ERL87627.1 KQ981909 KYN33188.1 GEZM01070974 JAV66153.1 GEZM01070975 JAV66152.1 GEZM01070976 JAV66151.1 GBYB01010781 JAG80548.1 KQ414666 KOC65093.1 AY739658 AAU85297.1 KQ434829 KZC07726.1 NNAY01001523 OXU23701.1 GEBQ01031210 JAT08767.1 GDKW01002950 JAI53645.1 QOIP01000002 RLU25421.1 DS235878 EEB19826.1 GL762889 EFZ20589.1 GEZM01070973 JAV66155.1 GDHC01014301 JAQ04328.1 GDHC01013584 JAQ05045.1 KQ971354 KYB26551.1 GECL01002210 JAP03914.1 GBGD01000828 JAC88061.1 ACPB03006006 ACPB03006007 ACPB03006008 GECZ01027105 JAS42664.1 GECZ01004576 JAS65193.1 GECU01033827 GECU01003848 JAS73879.1 JAT03859.1 DQ865256 ABI94039.1 DQ865250 ABI94033.1 DQ865258 ABI94041.1 MH368784 AZL49143.1 DQ865257 ABI94040.1 DQ865253 ABI94036.1 DQ865255 ABI94038.1 DQ865259 ABI94042.1 DQ865251 ABI94034.1 AY280847 AAQ16311.1 GEDC01030116 JAS07182.1 MWRG01001444 PRD34188.1 QCYY01001360 ROT78575.1 GDIQ01162353 JAK89372.1 GDIQ01039651 JAN55086.1 GDIP01050945 JAM52770.1 GDIP01199469 JAJ23933.1 GDIP01200779 JAJ22623.1 GDIP01201978 JAJ21424.1 GDIP01162922 JAJ60480.1 GDIQ01056438 JAN38299.1 GDIQ01180451 JAK71274.1 GDIP01236770 JAI86631.1 GDIP01209011 JAJ14391.1 GDIP01169965 JAJ53437.1 GDIQ01161010 JAK90715.1 GDIQ01196348 JAK55377.1 GDIQ01216041 JAK35684.1 GDIQ01059086 JAN35651.1 GDIQ01113921 JAL37805.1 GDIQ01180450 JAK71275.1 GDIQ01175789 JAK75936.1 GDIQ01175788 JAK75937.1 GDIQ01036395 JAN58342.1 GDIQ01239046 JAK12679.1 GDIP01194979 JAJ28423.1 GDIP01221774 JAJ01628.1 GDIP01220716 JAJ02686.1 GDIQ01235478 JAK16247.1 GDIP01153421 JAJ69981.1
Proteomes
UP000053268
UP000053240
UP000005204
UP000007151
UP000037510
UP000218220
+ More
UP000007755 UP000008237 UP000000311 UP000027135 UP000053097 UP000036403 UP000192223 UP000242457 UP000078542 UP000002358 UP000078540 UP000078492 UP000075809 UP000005205 UP000019118 UP000030742 UP000078541 UP000053825 UP000005203 UP000076502 UP000215335 UP000279307 UP000009046 UP000007266 UP000015103 UP000283509
UP000007755 UP000008237 UP000000311 UP000027135 UP000053097 UP000036403 UP000192223 UP000242457 UP000078542 UP000002358 UP000078540 UP000078492 UP000075809 UP000005205 UP000019118 UP000030742 UP000078541 UP000053825 UP000005203 UP000076502 UP000215335 UP000279307 UP000009046 UP000007266 UP000015103 UP000283509
PRIDE
Interpro
SUPFAM
SSF51206
SSF51206
Gene 3D
CDD
ProteinModelPortal
A0A194QEW2
A0A194RFP5
H9IX98
A0A212F2B3
A0A2H1V032
A0A0L7KPU8
+ More
O96777 A0A2A4K4L7 A0A2A4K4K4 F4W9U8 E2BZX9 E2A5U2 V9I8E8 A0A0C9RQN1 A0A067RJN2 V9IAA4 A0A026WCB0 A0A0J7P2B4 A0A1W4XCU8 V9I9T3 A0A2A3EAG6 Q6WL04 A0A1W4XCL5 A0A195CG79 K7INL3 A0A195BRD4 A0A1W4X249 A0A151JPH3 A0A151WKF2 A0A1B6K7N7 A0A158NZ19 A0A0T6BED1 Q5XQT5 N6SRH5 A0A195EY94 A0A1Y1KXL2 A0A1Y1KXP8 A0A1Y1L055 A0A0C9RTG4 A0A0L7R2N3 Q5XQT6 A0A088A929 A0A154P706 K7INL2 A0A232EZ95 A0A1B6KBD0 A0A0P4VVJ6 A0A3L8DY95 E0W2H0 E9IFR6 A0A1Y1L2G8 A0A146L7B4 A0A146LBW4 A0A139WFA2 A0A0V0G8Y1 A0A069DW42 T1HZI8 A0A1B6EXC7 A0A1B6GS55 A0A1B6HGY3 Q06IP4 Q06IQ0 Q06IP2 A0A3Q8TMV0 Q06IP3 Q06IP7 Q06IP5 Q06IP1 Q06IP9 Q6WL05 A0A1B6C0Y8 A0A2P6L6H5 A0A3R7SWG4 A0A0P5M0J4 A0A0P6GXS3 A0A0P5Z2H7 A0A0P5AGG2 A0A0P5ACY5 A0A0P5AA68 A0A0P5EAB5 A0A0P6FJJ4 A0A0N8BGC6 A0A0P4XWK1 A0A0P5ASL0 A0A0P5CIS0 A0A0P5ME34 A0A0P5JMZ8 A0A0P5IDY4 A0A0P6EU10 A0A0P5QHU2 A0A0P5MF30 A0A0P5LT45 A0A0P5MQ71 A0A0P6GA18 A0A0P5H259 A0A0P5ASP2 A0A0P4ZTE3 A0A0P4YXL4 A0A0P5GWF7 A0A0P5DZP8
O96777 A0A2A4K4L7 A0A2A4K4K4 F4W9U8 E2BZX9 E2A5U2 V9I8E8 A0A0C9RQN1 A0A067RJN2 V9IAA4 A0A026WCB0 A0A0J7P2B4 A0A1W4XCU8 V9I9T3 A0A2A3EAG6 Q6WL04 A0A1W4XCL5 A0A195CG79 K7INL3 A0A195BRD4 A0A1W4X249 A0A151JPH3 A0A151WKF2 A0A1B6K7N7 A0A158NZ19 A0A0T6BED1 Q5XQT5 N6SRH5 A0A195EY94 A0A1Y1KXL2 A0A1Y1KXP8 A0A1Y1L055 A0A0C9RTG4 A0A0L7R2N3 Q5XQT6 A0A088A929 A0A154P706 K7INL2 A0A232EZ95 A0A1B6KBD0 A0A0P4VVJ6 A0A3L8DY95 E0W2H0 E9IFR6 A0A1Y1L2G8 A0A146L7B4 A0A146LBW4 A0A139WFA2 A0A0V0G8Y1 A0A069DW42 T1HZI8 A0A1B6EXC7 A0A1B6GS55 A0A1B6HGY3 Q06IP4 Q06IQ0 Q06IP2 A0A3Q8TMV0 Q06IP3 Q06IP7 Q06IP5 Q06IP1 Q06IP9 Q6WL05 A0A1B6C0Y8 A0A2P6L6H5 A0A3R7SWG4 A0A0P5M0J4 A0A0P6GXS3 A0A0P5Z2H7 A0A0P5AGG2 A0A0P5ACY5 A0A0P5AA68 A0A0P5EAB5 A0A0P6FJJ4 A0A0N8BGC6 A0A0P4XWK1 A0A0P5ASL0 A0A0P5CIS0 A0A0P5ME34 A0A0P5JMZ8 A0A0P5IDY4 A0A0P6EU10 A0A0P5QHU2 A0A0P5MF30 A0A0P5LT45 A0A0P5MQ71 A0A0P6GA18 A0A0P5H259 A0A0P5ASP2 A0A0P4ZTE3 A0A0P4YXL4 A0A0P5GWF7 A0A0P5DZP8
PDB
5U6P
E-value=1.85761e-11,
Score=160
Ontologies
PANTHER
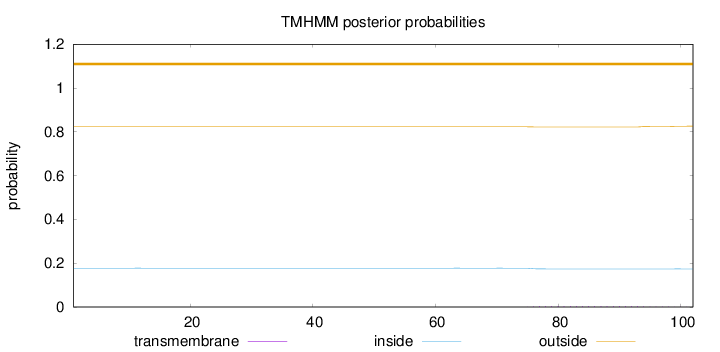
Topology
Length:
102
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09339
Exp number, first 60 AAs:
0.00035
Total prob of N-in:
0.17732
outside
1 - 102
Population Genetic Test Statistics
Pi
161.517436
Theta
164.993503
Tajima's D
0.038102
CLR
0.275073
CSRT
0.388130593470326
Interpretation
Uncertain
