Gene
KWMTBOMO11417
Pre Gene Modal
BGIBMGA001878
Annotation
PREDICTED:_fukutin-related_protein_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.133
Sequence
CDS
ATGTTCAGAAAAATCGTTTTCCACCGTTTATTACGCGGCCAGCAAGGATTTAAGCTGCAATTTTTCGTCTCGATCGCTGTTTTGGTGTTACTGTGCGTCTACCTATTTTCCCTGCTAACAGAAGTGACGTATAACAAAAGAGCAACAGATACTGAATCGGGCTGGCAGAAAAAGCCTTTGTCGTCGTTGGTAACTGTTGTGTTTCGGCAGTTCGAGGAAGCGGACAATGATGTTGCCGACAGCGTCCTGTCGTTCGTATCGGCCTTCCCGGATATGCAGGTGCTGGTCGTGTGCGAGAGGAGTCCGTACCCCCCGATCGAGCTTCCCCCTACCAACAGCTCTTATAGAAACGTAAAGATCTTGTCTCTCGACTACGAAATCGATTCCTGCCCGTACACCTCGAGTCCACTGAGTCCGTTTAATATCGAAACAGAGTATACACTTTTTGTGCCCGATTCAGTTCGGGTAAAACCACGGGTCTTGCATCAAGCTGTGATTGAAGCCAACGCCAATCCTTTACATGTCATAGCAATTGGAATCGGTTTCCATGAGATCAAATGTCAAAGAATCTGCTGGAACTACAGAGACTGGACGCTAAAGTATACTAAGGATTCAACAGGAACGATATGCGACGCCGTCACAGGTCCACATGCGCTATTAATTAAAACTGAATATCTGTCAAAGGTGCCAGATCCGATTGCGTTGCCGTTTCCAGAATCATTATATTTGCAGACATATGTGAAGATGGCCAAGGTACAAGTACTCTATCATACATTTGGACCGGGACGTAAAGTCTTAAAAACATTGCCAGAAAAACAAATAGTTGCTGAAAAAGTGAAAGAGCAGAGGTCGACATTCTATAAGAAATATAAAATGAAGATTGTTGTCGGAGAAGATGGAACAGTACACGAATATGGATGTGCTAAAGATACACCGAGATGTTTTCCAACAGTCAAAGGCAAACCTTCCTACGTCTACTCGGATAAACACACTCCGCCTTGCTGTCTCCGAAATATGCGTGAAGTTACGAAACACGTTCTGGATGTACTGGAGCAAGCTGGTGTCAGATGCTGGTTGGAAACATCATCTCTGCTTGGAGCTGTGGTAGATGGAAATGTCCTAGTCTGGGCAGATCATGTTGAGTTGGGGATTTACTCTTCAGACATAGATCGCGTACCGTATTTGCAGCGAGGTGGGACTGACGCTGACGGCTTTGTGTGGGAGCGCGCCACAAAAGGGCATTATTACAGCGTTTCACTCTCGGCGATTAACAAAGCGAATGTATTGATTCTACCGTTCACATCACAAAACGGTACCATGTGGCCTGCCGACTGGGTCCTGGCTCACCAAAGGGAGTTTCCGGAACGCCATTTGCATCCACTTGCCCAGATACCCTTCGCCTACGTACAAGCACCGGCTCCTAACGATGCACACGCTTTCCTGGACCTAAAATTGGGGCCGAATGCTGTCGAAAAATATAAAAAACTTGGAGCGAAACTTCTCTATCCTTAA
Protein
MFRKIVFHRLLRGQQGFKLQFFVSIAVLVLLCVYLFSLLTEVTYNKRATDTESGWQKKPLSSLVTVVFRQFEEADNDVADSVLSFVSAFPDMQVLVVCERSPYPPIELPPTNSSYRNVKILSLDYEIDSCPYTSSPLSPFNIETEYTLFVPDSVRVKPRVLHQAVIEANANPLHVIAIGIGFHEIKCQRICWNYRDWTLKYTKDSTGTICDAVTGPHALLIKTEYLSKVPDPIALPFPESLYLQTYVKMAKVQVLYHTFGPGRKVLKTLPEKQIVAEKVKEQRSTFYKKYKMKIVVGEDGTVHEYGCAKDTPRCFPTVKGKPSYVYSDKHTPPCCLRNMREVTKHVLDVLEQAGVRCWLETSSLLGAVVDGNVLVWADHVELGIYSSDIDRVPYLQRGGTDADGFVWERATKGHYYSVSLSAINKANVLILPFTSQNGTMWPADWVLAHQREFPERHLHPLAQIPFAYVQAPAPNDAHAFLDLKLGPNAVEKYKKLGAKLLYP
Summary
Uniprot
H9IX95
A0A2A4JLG9
A0A2H1W6T6
A0A212F985
A0A194QVN0
A0A194QEJ9
+ More
A0A154PK87 D6WSR7 A0A026W8W7 A0A0C9PVS1 A0A0L7R083 A0A195FXQ0 A0A195DRQ5 F4X2L3 A0A158NVA8 A0A0J7N1A7 E2B208 E2BE14 A0A087ZPC7 A0A2A3EIR9 A0A0N0BD59 A0A0L7LKF3 A0A2J7RBB6 K7INL0 A0A232EU57 A0A1B6LWN5 A0A067QXV6 A0A023FBF0 T1I9E1 A0A151IBU1 A0A026W805 A0A0A9WF93 A0A1B6ECW0 A0A1B6FU49 A0A3L8DK23 A0A2R5L9B7 A0A2P6KIL6 A0A0L7KWG8 A0A3N0YUV8 A0A087UDB5 W5NMH5 A0A1S3ITH8 A0A3B5B6R1 A0A3Q1HEZ0 A0A3Q3WM07 C3Y239 A0A3B4WB96 A0A3Q2WZQ3 A0A3P8QAH4 A0A3B4TLX8 A0A3P9CHH4 I3KXC8 A0A3Q1D4S0 A0A3Q0QNG6 A0A2U9B520 A0A3P8SU42 A0A3Q1I9E7 A0A3Q3D7R7 A0A3Q4ME77 A0A3Q3M001 A0A0P7X3Z9 A0A147BDC9 A0A1A7XZL2 A0A3B4GU84 A0A1A8JLR9 A0A3R7MQ86 A0A1A8NI14 A0A1A8MZ28 A0A1A8EQX4 A7RY81 A0A3Q3A4Q9 A0A1A7ZFM7 A0A3Q3IQR7 A0A3Q7UNT3 W5LTU2 A0A1V9XFD2 A0A1A8D8P0 A0A3P9QHH6 A0A3B5M067 A0A3Q2PMS3 A0A2U3VZM5 A0A087YR49 A0A3B3TI29 A0A146ZQC7 A0A146RUD7 A0A3B3YNX0 A0A1A8CSI2 M3ZPN4 A0A2Y9L2C9 A0A3Q3GNA5 G3P4C8 H2ZYF7 H3CQP5 A0A3Q7NR87 F1RM20 Q4KLJ4 Q4SPH1 H2U5W8 A0A3Q2D6W3 A0A3M6TED7
A0A154PK87 D6WSR7 A0A026W8W7 A0A0C9PVS1 A0A0L7R083 A0A195FXQ0 A0A195DRQ5 F4X2L3 A0A158NVA8 A0A0J7N1A7 E2B208 E2BE14 A0A087ZPC7 A0A2A3EIR9 A0A0N0BD59 A0A0L7LKF3 A0A2J7RBB6 K7INL0 A0A232EU57 A0A1B6LWN5 A0A067QXV6 A0A023FBF0 T1I9E1 A0A151IBU1 A0A026W805 A0A0A9WF93 A0A1B6ECW0 A0A1B6FU49 A0A3L8DK23 A0A2R5L9B7 A0A2P6KIL6 A0A0L7KWG8 A0A3N0YUV8 A0A087UDB5 W5NMH5 A0A1S3ITH8 A0A3B5B6R1 A0A3Q1HEZ0 A0A3Q3WM07 C3Y239 A0A3B4WB96 A0A3Q2WZQ3 A0A3P8QAH4 A0A3B4TLX8 A0A3P9CHH4 I3KXC8 A0A3Q1D4S0 A0A3Q0QNG6 A0A2U9B520 A0A3P8SU42 A0A3Q1I9E7 A0A3Q3D7R7 A0A3Q4ME77 A0A3Q3M001 A0A0P7X3Z9 A0A147BDC9 A0A1A7XZL2 A0A3B4GU84 A0A1A8JLR9 A0A3R7MQ86 A0A1A8NI14 A0A1A8MZ28 A0A1A8EQX4 A7RY81 A0A3Q3A4Q9 A0A1A7ZFM7 A0A3Q3IQR7 A0A3Q7UNT3 W5LTU2 A0A1V9XFD2 A0A1A8D8P0 A0A3P9QHH6 A0A3B5M067 A0A3Q2PMS3 A0A2U3VZM5 A0A087YR49 A0A3B3TI29 A0A146ZQC7 A0A146RUD7 A0A3B3YNX0 A0A1A8CSI2 M3ZPN4 A0A2Y9L2C9 A0A3Q3GNA5 G3P4C8 H2ZYF7 H3CQP5 A0A3Q7NR87 F1RM20 Q4KLJ4 Q4SPH1 H2U5W8 A0A3Q2D6W3 A0A3M6TED7
Pubmed
EMBL
BABH01018099
NWSH01001047
PCG72925.1
ODYU01006642
SOQ48666.1
AGBW02009615
+ More
OWR50316.1 KQ461073 KPJ09593.1 KQ459053 KPJ03963.1 KQ434943 KZC12266.1 KQ971354 EFA07689.2 KK107324 QOIP01000006 EZA52537.1 RLU21503.1 GBYB01005568 JAG75335.1 KQ414672 KOC64237.1 KQ981193 KYN45206.1 KQ980530 KYN15610.1 GL888584 EGI59338.1 ADTU01027053 LBMM01012057 KMQ86440.1 GL445031 EFN60264.1 GL447731 EFN86078.1 KZ288246 PBC31182.1 KQ435878 KOX69961.1 JTDY01000859 KOB75651.1 NEVH01005915 PNF38118.1 NNAY01002170 OXU21881.1 GEBQ01011927 JAT28050.1 KK853230 KDR09637.1 GBBI01000378 JAC18334.1 ACPB03016960 KQ978106 KYM96978.1 KK107347 EZA52237.1 GBHO01037538 GDHC01006486 JAG06066.1 JAQ12143.1 GEDC01001520 JAS35778.1 GECZ01016100 JAS53669.1 QOIP01000007 RLU20794.1 RLU20899.1 GGLE01001957 MBY06083.1 MWRG01011055 PRD26168.1 JTDY01005079 KOB67389.1 RJVU01024579 ROL49973.1 KK119315 KFM75354.1 AHAT01023928 GG666480 EEN65929.1 AERX01025081 CP026245 AWO98986.1 JARO02004298 KPP68758.1 GEGO01006638 JAR88766.1 HADW01011234 HADX01001365 SBP23597.1 HAED01020817 HAEE01000996 SBR21012.1 QCYY01002828 ROT67261.1 HAEH01002262 HAEI01013621 SBR68561.1 HAEF01020605 HAEG01007637 SBR61764.1 HAEB01002441 HAEC01014824 SBQ48968.1 DS469552 EDO43590.1 HADY01002808 HAEJ01009480 SBP41293.1 MNPL01012447 OQR72126.1 HAEA01002007 SBQ30487.1 AYCK01003474 GCES01017793 JAR68530.1 GCES01114662 JAQ71660.1 HADZ01017804 SBP81745.1 AFYH01111771 AEMK02000041 AC127887 BC099170 CH473979 AAH99170.1 EDM08306.1 CAAE01014537 CAF97461.1 RCHS01003787 RMX39742.1
OWR50316.1 KQ461073 KPJ09593.1 KQ459053 KPJ03963.1 KQ434943 KZC12266.1 KQ971354 EFA07689.2 KK107324 QOIP01000006 EZA52537.1 RLU21503.1 GBYB01005568 JAG75335.1 KQ414672 KOC64237.1 KQ981193 KYN45206.1 KQ980530 KYN15610.1 GL888584 EGI59338.1 ADTU01027053 LBMM01012057 KMQ86440.1 GL445031 EFN60264.1 GL447731 EFN86078.1 KZ288246 PBC31182.1 KQ435878 KOX69961.1 JTDY01000859 KOB75651.1 NEVH01005915 PNF38118.1 NNAY01002170 OXU21881.1 GEBQ01011927 JAT28050.1 KK853230 KDR09637.1 GBBI01000378 JAC18334.1 ACPB03016960 KQ978106 KYM96978.1 KK107347 EZA52237.1 GBHO01037538 GDHC01006486 JAG06066.1 JAQ12143.1 GEDC01001520 JAS35778.1 GECZ01016100 JAS53669.1 QOIP01000007 RLU20794.1 RLU20899.1 GGLE01001957 MBY06083.1 MWRG01011055 PRD26168.1 JTDY01005079 KOB67389.1 RJVU01024579 ROL49973.1 KK119315 KFM75354.1 AHAT01023928 GG666480 EEN65929.1 AERX01025081 CP026245 AWO98986.1 JARO02004298 KPP68758.1 GEGO01006638 JAR88766.1 HADW01011234 HADX01001365 SBP23597.1 HAED01020817 HAEE01000996 SBR21012.1 QCYY01002828 ROT67261.1 HAEH01002262 HAEI01013621 SBR68561.1 HAEF01020605 HAEG01007637 SBR61764.1 HAEB01002441 HAEC01014824 SBQ48968.1 DS469552 EDO43590.1 HADY01002808 HAEJ01009480 SBP41293.1 MNPL01012447 OQR72126.1 HAEA01002007 SBQ30487.1 AYCK01003474 GCES01017793 JAR68530.1 GCES01114662 JAQ71660.1 HADZ01017804 SBP81745.1 AFYH01111771 AEMK02000041 AC127887 BC099170 CH473979 AAH99170.1 EDM08306.1 CAAE01014537 CAF97461.1 RCHS01003787 RMX39742.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000076502
+ More
UP000007266 UP000053097 UP000279307 UP000053825 UP000078541 UP000078492 UP000007755 UP000005205 UP000036403 UP000000311 UP000008237 UP000005203 UP000242457 UP000053105 UP000037510 UP000235965 UP000002358 UP000215335 UP000027135 UP000015103 UP000078542 UP000054359 UP000018468 UP000085678 UP000261400 UP000257200 UP000261620 UP000001554 UP000261360 UP000264840 UP000265100 UP000261420 UP000265160 UP000005207 UP000257160 UP000261340 UP000246464 UP000265080 UP000265040 UP000264820 UP000261580 UP000261640 UP000034805 UP000261460 UP000283509 UP000001593 UP000264800 UP000261600 UP000286642 UP000018467 UP000192247 UP000242638 UP000261380 UP000265000 UP000245340 UP000028760 UP000261500 UP000261480 UP000002852 UP000248482 UP000261660 UP000007635 UP000008672 UP000007303 UP000286641 UP000008227 UP000002494 UP000005226 UP000265020 UP000275408
UP000007266 UP000053097 UP000279307 UP000053825 UP000078541 UP000078492 UP000007755 UP000005205 UP000036403 UP000000311 UP000008237 UP000005203 UP000242457 UP000053105 UP000037510 UP000235965 UP000002358 UP000215335 UP000027135 UP000015103 UP000078542 UP000054359 UP000018468 UP000085678 UP000261400 UP000257200 UP000261620 UP000001554 UP000261360 UP000264840 UP000265100 UP000261420 UP000265160 UP000005207 UP000257160 UP000261340 UP000246464 UP000265080 UP000265040 UP000264820 UP000261580 UP000261640 UP000034805 UP000261460 UP000283509 UP000001593 UP000264800 UP000261600 UP000286642 UP000018467 UP000192247 UP000242638 UP000261380 UP000265000 UP000245340 UP000028760 UP000261500 UP000261480 UP000002852 UP000248482 UP000261660 UP000007635 UP000008672 UP000007303 UP000286641 UP000008227 UP000002494 UP000005226 UP000265020 UP000275408
Pfam
PF04991 LicD
SUPFAM
SSF53448
SSF53448
ProteinModelPortal
H9IX95
A0A2A4JLG9
A0A2H1W6T6
A0A212F985
A0A194QVN0
A0A194QEJ9
+ More
A0A154PK87 D6WSR7 A0A026W8W7 A0A0C9PVS1 A0A0L7R083 A0A195FXQ0 A0A195DRQ5 F4X2L3 A0A158NVA8 A0A0J7N1A7 E2B208 E2BE14 A0A087ZPC7 A0A2A3EIR9 A0A0N0BD59 A0A0L7LKF3 A0A2J7RBB6 K7INL0 A0A232EU57 A0A1B6LWN5 A0A067QXV6 A0A023FBF0 T1I9E1 A0A151IBU1 A0A026W805 A0A0A9WF93 A0A1B6ECW0 A0A1B6FU49 A0A3L8DK23 A0A2R5L9B7 A0A2P6KIL6 A0A0L7KWG8 A0A3N0YUV8 A0A087UDB5 W5NMH5 A0A1S3ITH8 A0A3B5B6R1 A0A3Q1HEZ0 A0A3Q3WM07 C3Y239 A0A3B4WB96 A0A3Q2WZQ3 A0A3P8QAH4 A0A3B4TLX8 A0A3P9CHH4 I3KXC8 A0A3Q1D4S0 A0A3Q0QNG6 A0A2U9B520 A0A3P8SU42 A0A3Q1I9E7 A0A3Q3D7R7 A0A3Q4ME77 A0A3Q3M001 A0A0P7X3Z9 A0A147BDC9 A0A1A7XZL2 A0A3B4GU84 A0A1A8JLR9 A0A3R7MQ86 A0A1A8NI14 A0A1A8MZ28 A0A1A8EQX4 A7RY81 A0A3Q3A4Q9 A0A1A7ZFM7 A0A3Q3IQR7 A0A3Q7UNT3 W5LTU2 A0A1V9XFD2 A0A1A8D8P0 A0A3P9QHH6 A0A3B5M067 A0A3Q2PMS3 A0A2U3VZM5 A0A087YR49 A0A3B3TI29 A0A146ZQC7 A0A146RUD7 A0A3B3YNX0 A0A1A8CSI2 M3ZPN4 A0A2Y9L2C9 A0A3Q3GNA5 G3P4C8 H2ZYF7 H3CQP5 A0A3Q7NR87 F1RM20 Q4KLJ4 Q4SPH1 H2U5W8 A0A3Q2D6W3 A0A3M6TED7
A0A154PK87 D6WSR7 A0A026W8W7 A0A0C9PVS1 A0A0L7R083 A0A195FXQ0 A0A195DRQ5 F4X2L3 A0A158NVA8 A0A0J7N1A7 E2B208 E2BE14 A0A087ZPC7 A0A2A3EIR9 A0A0N0BD59 A0A0L7LKF3 A0A2J7RBB6 K7INL0 A0A232EU57 A0A1B6LWN5 A0A067QXV6 A0A023FBF0 T1I9E1 A0A151IBU1 A0A026W805 A0A0A9WF93 A0A1B6ECW0 A0A1B6FU49 A0A3L8DK23 A0A2R5L9B7 A0A2P6KIL6 A0A0L7KWG8 A0A3N0YUV8 A0A087UDB5 W5NMH5 A0A1S3ITH8 A0A3B5B6R1 A0A3Q1HEZ0 A0A3Q3WM07 C3Y239 A0A3B4WB96 A0A3Q2WZQ3 A0A3P8QAH4 A0A3B4TLX8 A0A3P9CHH4 I3KXC8 A0A3Q1D4S0 A0A3Q0QNG6 A0A2U9B520 A0A3P8SU42 A0A3Q1I9E7 A0A3Q3D7R7 A0A3Q4ME77 A0A3Q3M001 A0A0P7X3Z9 A0A147BDC9 A0A1A7XZL2 A0A3B4GU84 A0A1A8JLR9 A0A3R7MQ86 A0A1A8NI14 A0A1A8MZ28 A0A1A8EQX4 A7RY81 A0A3Q3A4Q9 A0A1A7ZFM7 A0A3Q3IQR7 A0A3Q7UNT3 W5LTU2 A0A1V9XFD2 A0A1A8D8P0 A0A3P9QHH6 A0A3B5M067 A0A3Q2PMS3 A0A2U3VZM5 A0A087YR49 A0A3B3TI29 A0A146ZQC7 A0A146RUD7 A0A3B3YNX0 A0A1A8CSI2 M3ZPN4 A0A2Y9L2C9 A0A3Q3GNA5 G3P4C8 H2ZYF7 H3CQP5 A0A3Q7NR87 F1RM20 Q4KLJ4 Q4SPH1 H2U5W8 A0A3Q2D6W3 A0A3M6TED7
Ontologies
PATHWAY
GO
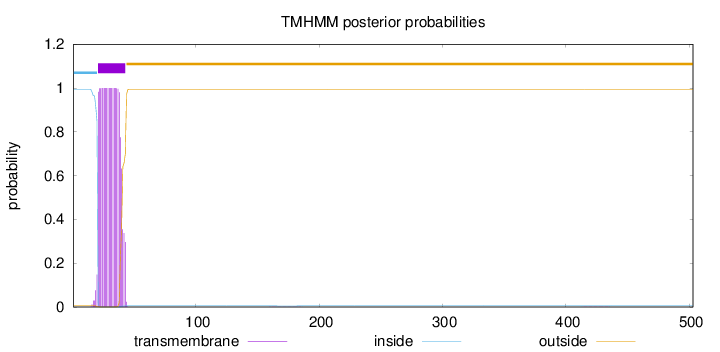
Topology
Length:
503
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.6872599999999
Exp number, first 60 AAs:
20.66704
Total prob of N-in:
0.99407
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 503
Population Genetic Test Statistics
Pi
190.432611
Theta
163.052526
Tajima's D
-0.626106
CLR
126.212265
CSRT
0.215789210539473
Interpretation
Uncertain
