Gene
KWMTBOMO11416 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001943
Annotation
PREDICTED:_ATPase_ASNA1_homolog_[Papilio_machaon]
Full name
ATPase ASNA1 homolog
+ More
Arsenical pump-driving ATPase
Arsenical pump-driving ATPase
Alternative Name
Arsenical pump-driving ATPase homolog
Arsenite-stimulated ATPase
Arsenite-stimulated ATPase
Location in the cell
Cytoplasmic Reliability : 3.117
Sequence
CDS
ATGGAAGATACTAAAGATTTTGAACCACTAGAACCTTCGTTGAAAAATGTTATCGATCAAAAGTCGTTAAGGTGGATTTTCGTTGGAGGGAAAGGCGGAGTTGGGAAAACTACGTGCAGTTGCAGTCTCGCAGTTCAGCTGTCGAAAGTTCGCGAATCTGTGTTAATCATATCAACTGATCCTGCCCACAATATCTCTGATGCATTCGACCAGAAATTTTCTAAAGTACCAACAAAGGTTAAAGGGTTTGACAACCTATTCGCTATGGAGATTGATCCGAATGTTGGATTAACAGAGCTGCCTGAAGAATATTTTGAAGGAGAATCTGAGGCTATGAGATTGGACAAGGGTGTCATGCAAGAGATTGTTGGAGCTTTTCCTGGTATTGATGAGGCTATGAGCTATGCTGAGGTGATGAAACTAGTAAAAGGCATGAACTTTAGTGCTGTAGTGTTCGATACTGCACCGACTGGACATACTCTCAGACTTCTATCATTCCCGCAAGTTGTAGAACGAGGTCTCGGCAAGCTGATGCGTTTGAAGTCAAAGGTTGCCCCCTTCATCAATCAAATTGCGTCACTGTTTGGACTAGCAGATTTCAACTCGGACATGTTCAGCAACAAAATGGATGAGATGTTATCAGTTATCAGACAAGTGAATGCACAGTTCAAAGACCCGAATCAAACTACATTTGTGTGTGTTTGTATCGCAGAGTTCCTATCGCTTTACGAAACCGAACGTTTAGTTCAAGAATTAACGCGATGTGGAATCGATACACATAATATTATAGTTAATCAGTTGCTGCTTCGTTCATCTGCTCCTTGTGAACTATGTGCAGCCAGGCATAAGGTCCAAGAAAAATATCTGGAGCAAATTGCAGACTTGTATGAAGACTTCCATGTGACAAAGCTACCCTTGCTGGAAAAGGAAGTCCGAGGAGCTTCCGCTGTAAATGCATTTTCAGAACTTCTCTTGAAGCCTTACGATCCTCCTACATCCTCCACGTGA
Protein
MEDTKDFEPLEPSLKNVIDQKSLRWIFVGGKGGVGKTTCSCSLAVQLSKVRESVLIISTDPAHNISDAFDQKFSKVPTKVKGFDNLFAMEIDPNVGLTELPEEYFEGESEAMRLDKGVMQEIVGAFPGIDEAMSYAEVMKLVKGMNFSAVVFDTAPTGHTLRLLSFPQVVERGLGKLMRLKSKVAPFINQIASLFGLADFNSDMFSNKMDEMLSVIRQVNAQFKDPNQTTFVCVCIAEFLSLYETERLVQELTRCGIDTHNIIVNQLLLRSSAPCELCAARHKVQEKYLEQIADLYEDFHVTKLPLLEKEVRGASAVNAFSELLLKPYDPPTSST
Summary
Description
ATPase required for the post-translational delivery of tail-anchored (TA) proteins to the endoplasmic reticulum. Recognizes and selectively binds the transmembrane domain of TA proteins in the cytosol. This complex then targets to the endoplasmic reticulum by membrane-bound receptors, where the tail-anchored protein is released for insertion. This process is regulated by ATP binding and hydrolysis. ATP binding drives the homodimer towards the closed dimer state, facilitating recognition of newly synthesized TA membrane proteins. ATP hydrolysis is required for insertion. Subsequently, the homodimer reverts towards the open dimer state, lowering its affinity for the membrane-bound receptor, and returning it to the cytosol to initiate a new round of targeting.
Subunit
Homodimer.
Similarity
Belongs to the arsA ATPase family.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Endoplasmic reticulum
Hydrolase
Metal-binding
Nucleotide-binding
Reference proteome
Transport
Zinc
Feature
chain ATPase ASNA1 homolog
Uniprot
H9IXG0
A0A2A4JMB3
A0A2H1W6L0
A0A212F9C0
S4PCC6
A0A0L7KW95
+ More
A0A0N1PHK1 A0A194QEW7 A0A0C9R9J4 A0A0J7KJI2 A0A154P793 E0VDM0 A0A026WTN5 A0A023EQ71 A0A2A3E513 V9IHT7 B0WEV5 A0A087ZY48 A0A067RLC5 A0A1B6G8W3 A0A1B6I1K2 A0A1B6DK67 A0A1Q3FCF8 A0A0M8ZVY9 K7IME4 A0A1B6MRI1 A0A0K8TNT1 Q16MG9 U5EIV3 A0A151WEM8 A0A0L7RCC5 A0A310SCD1 A0A182GNT7 W8BCD5 A0A1Y1NJ49 A0A0A1X261 A0A034VJ18 A0A195FGS3 E2BL31 A0A158NPA9 A0A195BEQ3 F4X362 A0A0C9RNL2 A0A151IVP1 A0A0K8U1B7 A0A232FDB4 A0A1W4UZX5 A0A151IL13 A0A0J9TVE2 B4QEC4 B4P1R6 A0A1I8MPI4 A0A1A9VQH0 A0A336LP49 D3TN99 A0A1I8P9Y2 A0A1L8DFI2 B3N9X2 Q7JWD3 A0A1L8DFB0 A0A1A9Z569 A0A1A9YJE2 A0A1B0C0I7 Q28YJ2 B4H8J5 B3MHB7 A0A1B0CAG5 A0A2M4AFJ3 A0A2J7QQB0 A0A182JGD8 A0A084WED0 B4J4F6 A0A3B0JXA3 E2AIS3 B4KTG7 B4LN33 W5JQD0 A0A2M4BSP7 A0A2M4BSM7 A0A2M4BSP8 A0A0M4EVN8 A0A2M3Z582 B4N645 A0A2M3ZH77 A0A1A9WQH6 A0A182T3X5 A0A023EP66 T1DGA5 A0A182I2D3 A0A182FHD5 A0A182XMK5 A0A182JVQ8 A0A182UVV4 A0A182KQ73 Q5TRE7 D2A135 A0A182NJC5 A0A182R4J5 A0A182PUZ3 A0A0P5RW25
A0A0N1PHK1 A0A194QEW7 A0A0C9R9J4 A0A0J7KJI2 A0A154P793 E0VDM0 A0A026WTN5 A0A023EQ71 A0A2A3E513 V9IHT7 B0WEV5 A0A087ZY48 A0A067RLC5 A0A1B6G8W3 A0A1B6I1K2 A0A1B6DK67 A0A1Q3FCF8 A0A0M8ZVY9 K7IME4 A0A1B6MRI1 A0A0K8TNT1 Q16MG9 U5EIV3 A0A151WEM8 A0A0L7RCC5 A0A310SCD1 A0A182GNT7 W8BCD5 A0A1Y1NJ49 A0A0A1X261 A0A034VJ18 A0A195FGS3 E2BL31 A0A158NPA9 A0A195BEQ3 F4X362 A0A0C9RNL2 A0A151IVP1 A0A0K8U1B7 A0A232FDB4 A0A1W4UZX5 A0A151IL13 A0A0J9TVE2 B4QEC4 B4P1R6 A0A1I8MPI4 A0A1A9VQH0 A0A336LP49 D3TN99 A0A1I8P9Y2 A0A1L8DFI2 B3N9X2 Q7JWD3 A0A1L8DFB0 A0A1A9Z569 A0A1A9YJE2 A0A1B0C0I7 Q28YJ2 B4H8J5 B3MHB7 A0A1B0CAG5 A0A2M4AFJ3 A0A2J7QQB0 A0A182JGD8 A0A084WED0 B4J4F6 A0A3B0JXA3 E2AIS3 B4KTG7 B4LN33 W5JQD0 A0A2M4BSP7 A0A2M4BSM7 A0A2M4BSP8 A0A0M4EVN8 A0A2M3Z582 B4N645 A0A2M3ZH77 A0A1A9WQH6 A0A182T3X5 A0A023EP66 T1DGA5 A0A182I2D3 A0A182FHD5 A0A182XMK5 A0A182JVQ8 A0A182UVV4 A0A182KQ73 Q5TRE7 D2A135 A0A182NJC5 A0A182R4J5 A0A182PUZ3 A0A0P5RW25
EC Number
3.6.-.-
Pubmed
19121390
22118469
23622113
26227816
26354079
20566863
+ More
24508170 30249741 24945155 24845553 20075255 26369729 17510324 26483478 24495485 28004739 25830018 25348373 20798317 21347285 21719571 28648823 22936249 17994087 25315136 20353571 10731132 12537572 12537569 15632085 24438588 20920257 23761445 20966253 12364791 18362917 19820115
24508170 30249741 24945155 24845553 20075255 26369729 17510324 26483478 24495485 28004739 25830018 25348373 20798317 21347285 21719571 28648823 22936249 17994087 25315136 20353571 10731132 12537572 12537569 15632085 24438588 20920257 23761445 20966253 12364791 18362917 19820115
EMBL
BABH01018100
NWSH01001047
PCG72926.1
ODYU01006642
SOQ48667.1
AGBW02009615
+ More
OWR50317.1 GAIX01005502 JAA87058.1 JTDY01005079 KOB67390.1 KQ460772 KPJ12479.1 KQ459053 KPJ03964.1 GBYB01013084 JAG82851.1 LBMM01006692 KMQ90391.1 KQ434829 KZC07747.1 DS235082 EEB11476.1 KK107106 QOIP01000002 EZA59425.1 RLU25731.1 GAPW01002393 JAC11205.1 KZ288369 PBC26778.1 JR046608 AEY60222.1 DS231911 KK852607 KDR20350.1 GECZ01010892 JAS58877.1 GECU01026898 JAS80808.1 GEDC01011231 JAS26067.1 GFDL01009853 JAV25192.1 KQ435847 KOX71146.1 GEBQ01001482 JAT38495.1 GDAI01001579 JAI16024.1 CH477862 CH477728 GANO01002482 JAB57389.1 KQ983238 KYQ46245.1 KQ414616 KOC68450.1 KQ769179 OAD52923.1 JXUM01077077 JXUM01080023 KQ563159 KQ562985 KXJ74379.1 KXJ74707.1 GAMC01011888 GAMC01011887 JAB94668.1 GEZM01001382 GEZM01001381 GEZM01001380 GEZM01001379 GEZM01001378 GEZM01001377 JAV97901.1 GBXI01008868 JAD05424.1 GAKP01015696 JAC43256.1 KQ981606 KYN39576.1 GL448906 EFN83600.1 ADTU01000325 KQ976509 KYM82689.1 GL888609 EGI59043.1 GBYB01009995 JAG79762.1 KQ980895 KYN11741.1 GDHF01032184 JAI20130.1 NNAY01000431 OXU28439.1 KQ977151 KYN05294.1 CM002911 KMY92015.1 CM000362 CM000157 UFQT01000091 UFQT01000704 SSX19565.1 SSX26667.1 EZ422901 ADD19177.1 GFDF01008947 JAV05137.1 CH954177 AE013599 AY070974 AY113636 BT044579 GFDF01008948 JAV05136.1 JXJN01023642 CM000071 CH479223 CH902619 AJWK01003749 GGFK01006212 MBW39533.1 NEVH01012087 PNF30769.1 ATLV01023201 KE525341 KFB48574.1 CH916367 OUUW01000001 SPP75718.1 GL439874 EFN66662.1 CH933808 CH940648 ADMH02000830 ETN64944.1 GGFJ01006964 MBW56105.1 GGFJ01006944 MBW56085.1 GGFJ01006949 MBW56090.1 CP012524 ALC41720.1 GGFM01002926 MBW23677.1 CH964154 GGFM01007100 MBW27851.1 GAPW01002822 JAC10776.1 GAMD01002836 JAA98754.1 APCN01000146 AAAB01008960 KQ971338 EFA01588.1 GDIQ01104703 JAL47023.1
OWR50317.1 GAIX01005502 JAA87058.1 JTDY01005079 KOB67390.1 KQ460772 KPJ12479.1 KQ459053 KPJ03964.1 GBYB01013084 JAG82851.1 LBMM01006692 KMQ90391.1 KQ434829 KZC07747.1 DS235082 EEB11476.1 KK107106 QOIP01000002 EZA59425.1 RLU25731.1 GAPW01002393 JAC11205.1 KZ288369 PBC26778.1 JR046608 AEY60222.1 DS231911 KK852607 KDR20350.1 GECZ01010892 JAS58877.1 GECU01026898 JAS80808.1 GEDC01011231 JAS26067.1 GFDL01009853 JAV25192.1 KQ435847 KOX71146.1 GEBQ01001482 JAT38495.1 GDAI01001579 JAI16024.1 CH477862 CH477728 GANO01002482 JAB57389.1 KQ983238 KYQ46245.1 KQ414616 KOC68450.1 KQ769179 OAD52923.1 JXUM01077077 JXUM01080023 KQ563159 KQ562985 KXJ74379.1 KXJ74707.1 GAMC01011888 GAMC01011887 JAB94668.1 GEZM01001382 GEZM01001381 GEZM01001380 GEZM01001379 GEZM01001378 GEZM01001377 JAV97901.1 GBXI01008868 JAD05424.1 GAKP01015696 JAC43256.1 KQ981606 KYN39576.1 GL448906 EFN83600.1 ADTU01000325 KQ976509 KYM82689.1 GL888609 EGI59043.1 GBYB01009995 JAG79762.1 KQ980895 KYN11741.1 GDHF01032184 JAI20130.1 NNAY01000431 OXU28439.1 KQ977151 KYN05294.1 CM002911 KMY92015.1 CM000362 CM000157 UFQT01000091 UFQT01000704 SSX19565.1 SSX26667.1 EZ422901 ADD19177.1 GFDF01008947 JAV05137.1 CH954177 AE013599 AY070974 AY113636 BT044579 GFDF01008948 JAV05136.1 JXJN01023642 CM000071 CH479223 CH902619 AJWK01003749 GGFK01006212 MBW39533.1 NEVH01012087 PNF30769.1 ATLV01023201 KE525341 KFB48574.1 CH916367 OUUW01000001 SPP75718.1 GL439874 EFN66662.1 CH933808 CH940648 ADMH02000830 ETN64944.1 GGFJ01006964 MBW56105.1 GGFJ01006944 MBW56085.1 GGFJ01006949 MBW56090.1 CP012524 ALC41720.1 GGFM01002926 MBW23677.1 CH964154 GGFM01007100 MBW27851.1 GAPW01002822 JAC10776.1 GAMD01002836 JAA98754.1 APCN01000146 AAAB01008960 KQ971338 EFA01588.1 GDIQ01104703 JAL47023.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
+ More
UP000036403 UP000076502 UP000009046 UP000053097 UP000279307 UP000242457 UP000002320 UP000005203 UP000027135 UP000053105 UP000002358 UP000008820 UP000075809 UP000053825 UP000069940 UP000249989 UP000078541 UP000008237 UP000005205 UP000078540 UP000007755 UP000078492 UP000215335 UP000192221 UP000078542 UP000000304 UP000002282 UP000095301 UP000078200 UP000095300 UP000008711 UP000000803 UP000092445 UP000092443 UP000092460 UP000001819 UP000008744 UP000007801 UP000092461 UP000235965 UP000075880 UP000030765 UP000001070 UP000268350 UP000000311 UP000009192 UP000008792 UP000000673 UP000092553 UP000007798 UP000091820 UP000075901 UP000075840 UP000069272 UP000076407 UP000075881 UP000075903 UP000075882 UP000007062 UP000007266 UP000075884 UP000075900 UP000075885
UP000036403 UP000076502 UP000009046 UP000053097 UP000279307 UP000242457 UP000002320 UP000005203 UP000027135 UP000053105 UP000002358 UP000008820 UP000075809 UP000053825 UP000069940 UP000249989 UP000078541 UP000008237 UP000005205 UP000078540 UP000007755 UP000078492 UP000215335 UP000192221 UP000078542 UP000000304 UP000002282 UP000095301 UP000078200 UP000095300 UP000008711 UP000000803 UP000092445 UP000092443 UP000092460 UP000001819 UP000008744 UP000007801 UP000092461 UP000235965 UP000075880 UP000030765 UP000001070 UP000268350 UP000000311 UP000009192 UP000008792 UP000000673 UP000092553 UP000007798 UP000091820 UP000075901 UP000075840 UP000069272 UP000076407 UP000075881 UP000075903 UP000075882 UP000007062 UP000007266 UP000075884 UP000075900 UP000075885
Pfam
PF02374 ArsA_ATPase
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9IXG0
A0A2A4JMB3
A0A2H1W6L0
A0A212F9C0
S4PCC6
A0A0L7KW95
+ More
A0A0N1PHK1 A0A194QEW7 A0A0C9R9J4 A0A0J7KJI2 A0A154P793 E0VDM0 A0A026WTN5 A0A023EQ71 A0A2A3E513 V9IHT7 B0WEV5 A0A087ZY48 A0A067RLC5 A0A1B6G8W3 A0A1B6I1K2 A0A1B6DK67 A0A1Q3FCF8 A0A0M8ZVY9 K7IME4 A0A1B6MRI1 A0A0K8TNT1 Q16MG9 U5EIV3 A0A151WEM8 A0A0L7RCC5 A0A310SCD1 A0A182GNT7 W8BCD5 A0A1Y1NJ49 A0A0A1X261 A0A034VJ18 A0A195FGS3 E2BL31 A0A158NPA9 A0A195BEQ3 F4X362 A0A0C9RNL2 A0A151IVP1 A0A0K8U1B7 A0A232FDB4 A0A1W4UZX5 A0A151IL13 A0A0J9TVE2 B4QEC4 B4P1R6 A0A1I8MPI4 A0A1A9VQH0 A0A336LP49 D3TN99 A0A1I8P9Y2 A0A1L8DFI2 B3N9X2 Q7JWD3 A0A1L8DFB0 A0A1A9Z569 A0A1A9YJE2 A0A1B0C0I7 Q28YJ2 B4H8J5 B3MHB7 A0A1B0CAG5 A0A2M4AFJ3 A0A2J7QQB0 A0A182JGD8 A0A084WED0 B4J4F6 A0A3B0JXA3 E2AIS3 B4KTG7 B4LN33 W5JQD0 A0A2M4BSP7 A0A2M4BSM7 A0A2M4BSP8 A0A0M4EVN8 A0A2M3Z582 B4N645 A0A2M3ZH77 A0A1A9WQH6 A0A182T3X5 A0A023EP66 T1DGA5 A0A182I2D3 A0A182FHD5 A0A182XMK5 A0A182JVQ8 A0A182UVV4 A0A182KQ73 Q5TRE7 D2A135 A0A182NJC5 A0A182R4J5 A0A182PUZ3 A0A0P5RW25
A0A0N1PHK1 A0A194QEW7 A0A0C9R9J4 A0A0J7KJI2 A0A154P793 E0VDM0 A0A026WTN5 A0A023EQ71 A0A2A3E513 V9IHT7 B0WEV5 A0A087ZY48 A0A067RLC5 A0A1B6G8W3 A0A1B6I1K2 A0A1B6DK67 A0A1Q3FCF8 A0A0M8ZVY9 K7IME4 A0A1B6MRI1 A0A0K8TNT1 Q16MG9 U5EIV3 A0A151WEM8 A0A0L7RCC5 A0A310SCD1 A0A182GNT7 W8BCD5 A0A1Y1NJ49 A0A0A1X261 A0A034VJ18 A0A195FGS3 E2BL31 A0A158NPA9 A0A195BEQ3 F4X362 A0A0C9RNL2 A0A151IVP1 A0A0K8U1B7 A0A232FDB4 A0A1W4UZX5 A0A151IL13 A0A0J9TVE2 B4QEC4 B4P1R6 A0A1I8MPI4 A0A1A9VQH0 A0A336LP49 D3TN99 A0A1I8P9Y2 A0A1L8DFI2 B3N9X2 Q7JWD3 A0A1L8DFB0 A0A1A9Z569 A0A1A9YJE2 A0A1B0C0I7 Q28YJ2 B4H8J5 B3MHB7 A0A1B0CAG5 A0A2M4AFJ3 A0A2J7QQB0 A0A182JGD8 A0A084WED0 B4J4F6 A0A3B0JXA3 E2AIS3 B4KTG7 B4LN33 W5JQD0 A0A2M4BSP7 A0A2M4BSM7 A0A2M4BSP8 A0A0M4EVN8 A0A2M3Z582 B4N645 A0A2M3ZH77 A0A1A9WQH6 A0A182T3X5 A0A023EP66 T1DGA5 A0A182I2D3 A0A182FHD5 A0A182XMK5 A0A182JVQ8 A0A182UVV4 A0A182KQ73 Q5TRE7 D2A135 A0A182NJC5 A0A182R4J5 A0A182PUZ3 A0A0P5RW25
PDB
2WOO
E-value=5.47894e-98,
Score=912
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Endoplasmic reticulum
Endoplasmic reticulum
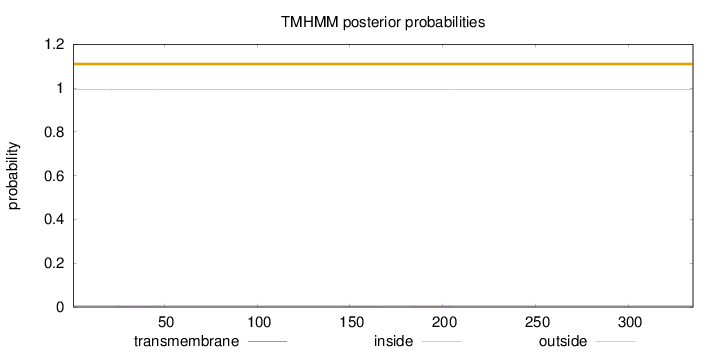
Length:
335
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0359
Exp number, first 60 AAs:
0.01393
Total prob of N-in:
0.00814
outside
1 - 335
Population Genetic Test Statistics
Pi
223.851998
Theta
188.620741
Tajima's D
0.572294
CLR
0.003999
CSRT
0.534073296335183
Interpretation
Possibly Positive selection
