Gene
KWMTBOMO11415
Pre Gene Modal
BGIBMGA001877
Annotation
PREDICTED:_GPI_ethanolamine_phosphate_transferase_3_isoform_X2_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.873
Sequence
CDS
ATGAGTGATATTACAGAATGTCAGCATTTGGAAACATTCGACTGCAGTGGACGAGAAAGAGGGAACTCAAGTATTGAAGAATCGTGCACATTGGATGAGAAAATAAAACAAATTTTAAGCGTGACCGGCTCTCCGTTAATATGCGCTCCATCCCATGGAAGAGTTGTCTTTATTTTAGTAGACGCATTGAGGTATGACTTCACAGAATATGATGACAAACTAGAAAAGCCTTTACCCTACCAGAACAGGCTGCCAGTGATGCAAAGGACTCTTGAATTATGCCCCGATTGTGTGCGCCTGTTTCGTTTCATCGCAGACCCACCAACAACCACTCTTCAACGTGTTAAAGCTCTTGTAACTGGTTCACTTCCCACTTTCATCGATGCAAGCTCTAATTTTGCAGCTATGGAGTTGCAAGAAGATAACATCATAGATCAGGTGGTGAATGCTGGACATCATGCAGTATTATTAGGAGATGACACTTGGTCGAGACTCATGCCAAGAAGATGGTTCCGGGCTCATACAATGTACTCATTCCACACTTGGGACCTTGACACTGTAGACATAGAAGTTGATTCAAAAATATATGATGAACTCAAAAAAGACGATTGGGATCTACTGGTAGCACATTATTTAGGAGTTGATCATGCCGGTCACAGGTATGGTCCAAACCATTCAGAAATGAAACGTAAATTGGATGAAACTAATGCAAGAATTGAGAAAATCATCAAAATTATACCTAAAGATGTCATCCTGTATGTTGTTGGAGACCACGGAATGACTGAATCAGGAGATCATGGTGGAGAGAGTAAGGCAGAAAGAACAGCTGCAATGTTTGCATATCGTGGTGCTGGATTTGGTGGTCAAAGCCCAGATATACAAACAGGTAGAGAAGTTGAACAGACTGATTTAGCGCCAACAATGAGTGCTGCTTTTGGTCGTCCACCGCCCGCACCATCCCTTGGCAACATTCTGTTTCCAGTACTGCCAAAAATGACCGTAGCCGAGACGCTACTGCATTTAACTAACAGCCTCAAGCAGGTTTCTCAATACTTAGTCCGTTACGGTGAAGAATCTCAACAGGTATCTTTAGATCGATTGGCGCATCTTATTAACGCGACAAGAGAACAGATAGAGAAAGCTGCAACGGTAAAGACTGAAGATGATCTCTCTATTTACGTTTCGAATGTTAGATTGTTAATGGACAACGTGCGCATAGTTTTTCGGGAAGTTTGGGTTGAGTTTGACACAGTTTCAATGCTTAGAGCACTTTTGTTACTTATTCTGGTGATATTTTTTAATTGGTTGATTGTTGAAGGCATACCCATAGAACGTCTTCCAAATATTTTTGCAAGTACTTTCGTTTCTTGTGGCCTGATATCAATGGCTATTTGCGTAAGCGTATGTTACACTGTCTTTCATTTTGAACTGTTAGAAGATGTTCATCATGGCGTTATTTTAAGCACGGGCTTAATATCTAGTGCGTTGACTTGCGTTTTGGTGATCATGCATTGGGACGGTATTTCACAACGGTGGTATGAAGGGAGATCGCCAATTTACGAGAGATTCTCCCGTGGCGCCTTAATGGCCTCAGCTGCTGTGTTATTATCTAATAGTTACATTATAGAAGAAGGAGCTGAATTATCTTTTCTAGCGTTATCGGTATTGGGCACGATTGCTTGGAATATTGGGACGATTAAGGCTTTCACTTTGTGGGTGGGTTTCGGAGCCACATTAGTAATTAGTAGATCGTATAGGGGATGTAGAGAGGAACAAGGAGATTGTTGGACTTCAATAGGTGTTGGCTCAACTGGACAGGCTTCCCGGACAGCGTTAGTAATGGCTTTAGGTTCAATGGCAGCTGTGGTTGCAATAGCGAGACGACATGTCGGTTGGCGAGGTCATGGTGTCGTCTTGGCTGGTTTGTTTGCATGCGCCCATTGGGCCCTCGGTTGGGGGGCTTTAGGTTCGCCCAGCAGATCAAGACAACTAGCCCGTGGATCATGGCTGATACTCGGATCGATGTTCGTTTTGCTTTGGAAAAGGGAAAGGTTTGGAGCAATACTGCCGTTAATTGTGTGCAGTTTACTCTTCTTCATTGCCAACGCCCTCGTTCTTGGTGCGTCTTTTGCTCCGTCTGCCGCTTTAGGACTATTGGCTGGATATTTGGCTCTTAACGTTGTTTCCATGATGAAGAATGGGAACTCTAAATTCTCATTATCAACTCGATGCAGCTCGAGTGTGGCGTGCATGTGGGCTCTGCTGGCGTCTTACCAATTTTATGGAACGGGCCACCATGCCACGTTGGGCCACGTACGATGGGCCCCGGCCTTCCACGCCGGTGATCCCAACTACCTCCCTATTGGACCTTTTGTCACTGGTGCTCTTGTCTCCTTGGAATTATTTGGTATGCCATTTATGTTCGGTGCCGCGGTTCCCTTGCTCGCGCTGTGGGGACGCAACGGTGCGGCCGCGGCGGGTCCCAGGACACAGATGGCGGCTGTCTTCACATTGTGCCTCAAGTTTGGATTGTGTTTTGCTGTCAGAGTGTTCATGAGTGCATTATCGGCTACAATACACTGTCGCCACTTAATGATATGGGGCGTGTTCACACCGAAGCTCTTGTTCGAGAGTGGGGCTTGTGCGGCCGCCCTGCTCGGTACCGTGGTGGGGGCCACGCTGACGGCCTGGCACGTGCCTACACAGATTAAGCCCAGTTGA
Protein
MSDITECQHLETFDCSGRERGNSSIEESCTLDEKIKQILSVTGSPLICAPSHGRVVFILVDALRYDFTEYDDKLEKPLPYQNRLPVMQRTLELCPDCVRLFRFIADPPTTTLQRVKALVTGSLPTFIDASSNFAAMELQEDNIIDQVVNAGHHAVLLGDDTWSRLMPRRWFRAHTMYSFHTWDLDTVDIEVDSKIYDELKKDDWDLLVAHYLGVDHAGHRYGPNHSEMKRKLDETNARIEKIIKIIPKDVILYVVGDHGMTESGDHGGESKAERTAAMFAYRGAGFGGQSPDIQTGREVEQTDLAPTMSAAFGRPPPAPSLGNILFPVLPKMTVAETLLHLTNSLKQVSQYLVRYGEESQQVSLDRLAHLINATREQIEKAATVKTEDDLSIYVSNVRLLMDNVRIVFREVWVEFDTVSMLRALLLLILVIFFNWLIVEGIPIERLPNIFASTFVSCGLISMAICVSVCYTVFHFELLEDVHHGVILSTGLISSALTCVLVIMHWDGISQRWYEGRSPIYERFSRGALMASAAVLLSNSYIIEEGAELSFLALSVLGTIAWNIGTIKAFTLWVGFGATLVISRSYRGCREEQGDCWTSIGVGSTGQASRTALVMALGSMAAVVAIARRHVGWRGHGVVLAGLFACAHWALGWGALGSPSRSRQLARGSWLILGSMFVLLWKRERFGAILPLIVCSLLFFIANALVLGASFAPSAALGLLAGYLALNVVSMMKNGNSKFSLSTRCSSSVACMWALLASYQFYGTGHHATLGHVRWAPAFHAGDPNYLPIGPFVTGALVSLELFGMPFMFGAAVPLLALWGRNGAAAAGPRTQMAAVFTLCLKFGLCFAVRVFMSALSATIHCRHLMIWGVFTPKLLFESGACAAALLGTVVGATLTAWHVPTQIKPS
Summary
Uniprot
H9IX94
A0A3S2TS99
A0A2A4JLJ3
A0A2A4JMI4
A0A194QG85
A0A2H1W6I0
+ More
A0A0N1I8I2 A0A212F9B4 A0A0L7LJR7 A0A139WFE5 E2BQR6 A0A182JE70 A0A2J7PJ72 E0VLL7 A0A088A583 A0A0C9PZY4 A0A1B6J892 A0A1S4F1U4 A0A1B6CPU1 A0A2A3ESJ3 A0A034WNW5 A0A084WKG3 A0A182G324 A0A0A1WGI8 A0A182G332 N6SW48 A0A0K8UNR6 Q17I41 A0A0L0CBV3 W5JUW6 A0A182YKC5 A0A1B0CLM7 A0A0L7QXF1 B4KM92 W8BEH6 A0A1W4U582 A0A182RRX3 A0A1L8DJZ9 B3MIW2 A0A1I8PKM7 A0A182TE79 A0A182UT63 A0A182LLF0 A0A182I126 A0A1I8N2W4 A0A1A9XDF0 A0A182X8Q0 Q7YU86 A0A1B0AMK9 A0A182MCH6 A1ZB84 B3NMT0 A0A1A9UR96 Q7Q5Q3 A0A1J1J8N9 A0A2R7W8K5
A0A0N1I8I2 A0A212F9B4 A0A0L7LJR7 A0A139WFE5 E2BQR6 A0A182JE70 A0A2J7PJ72 E0VLL7 A0A088A583 A0A0C9PZY4 A0A1B6J892 A0A1S4F1U4 A0A1B6CPU1 A0A2A3ESJ3 A0A034WNW5 A0A084WKG3 A0A182G324 A0A0A1WGI8 A0A182G332 N6SW48 A0A0K8UNR6 Q17I41 A0A0L0CBV3 W5JUW6 A0A182YKC5 A0A1B0CLM7 A0A0L7QXF1 B4KM92 W8BEH6 A0A1W4U582 A0A182RRX3 A0A1L8DJZ9 B3MIW2 A0A1I8PKM7 A0A182TE79 A0A182UT63 A0A182LLF0 A0A182I126 A0A1I8N2W4 A0A1A9XDF0 A0A182X8Q0 Q7YU86 A0A1B0AMK9 A0A182MCH6 A1ZB84 B3NMT0 A0A1A9UR96 Q7Q5Q3 A0A1J1J8N9 A0A2R7W8K5
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
20798317 20566863 17510324 25348373 24438588 26483478 25830018 23537049 26108605 20920257 23761445 25244985 17994087 24495485 20966253 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791
20798317 20566863 17510324 25348373 24438588 26483478 25830018 23537049 26108605 20920257 23761445 25244985 17994087 24495485 20966253 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791
EMBL
BABH01018100
RSAL01000010
RVE53739.1
NWSH01001047
PCG72927.1
PCG72928.1
+ More
KQ459053 KPJ03965.1 ODYU01006642 SOQ48668.1 KQ460772 KPJ12478.1 AGBW02009615 OWR50318.1 JTDY01000859 KOB75650.1 KQ971352 KYB26700.1 GL449799 EFN81941.1 NEVH01024950 PNF16390.1 DS235274 EEB14273.1 GBYB01007127 GBYB01007128 JAG76894.1 JAG76895.1 GECU01012309 JAS95397.1 GEDC01021838 JAS15460.1 KZ288191 PBC34484.1 GAKP01002613 JAC56339.1 ATLV01024119 KE525349 KFB50707.1 JXUM01040307 KQ561240 KXJ79255.1 GBXI01016769 JAC97522.1 JXUM01040322 KXJ79263.1 APGK01054078 APGK01054079 KB741247 KB632281 ENN71984.1 ERL91321.1 GDHF01023997 JAI28317.1 CH477243 EAT46318.1 JRES01000729 KNC28924.1 ADMH02000069 ETN67916.1 AJWK01017457 KQ414705 KOC63236.1 CH933808 EDW08756.1 GAMC01014959 JAB91596.1 GFDF01007301 JAV06783.1 CH902619 EDV36022.1 APCN01000024 BT009934 AAQ22403.1 JXJN01000495 AXCM01005995 AE013599 BT150350 AAF57689.1 AGW52159.1 CH954179 EDV55084.1 AAAB01008960 EAA10751.3 CVRI01000075 CRL08752.1 KK854464 PTY16063.1
KQ459053 KPJ03965.1 ODYU01006642 SOQ48668.1 KQ460772 KPJ12478.1 AGBW02009615 OWR50318.1 JTDY01000859 KOB75650.1 KQ971352 KYB26700.1 GL449799 EFN81941.1 NEVH01024950 PNF16390.1 DS235274 EEB14273.1 GBYB01007127 GBYB01007128 JAG76894.1 JAG76895.1 GECU01012309 JAS95397.1 GEDC01021838 JAS15460.1 KZ288191 PBC34484.1 GAKP01002613 JAC56339.1 ATLV01024119 KE525349 KFB50707.1 JXUM01040307 KQ561240 KXJ79255.1 GBXI01016769 JAC97522.1 JXUM01040322 KXJ79263.1 APGK01054078 APGK01054079 KB741247 KB632281 ENN71984.1 ERL91321.1 GDHF01023997 JAI28317.1 CH477243 EAT46318.1 JRES01000729 KNC28924.1 ADMH02000069 ETN67916.1 AJWK01017457 KQ414705 KOC63236.1 CH933808 EDW08756.1 GAMC01014959 JAB91596.1 GFDF01007301 JAV06783.1 CH902619 EDV36022.1 APCN01000024 BT009934 AAQ22403.1 JXJN01000495 AXCM01005995 AE013599 BT150350 AAF57689.1 AGW52159.1 CH954179 EDV55084.1 AAAB01008960 EAA10751.3 CVRI01000075 CRL08752.1 KK854464 PTY16063.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000007266 UP000008237 UP000075880 UP000235965 UP000009046 UP000005203 UP000242457 UP000030765 UP000069940 UP000249989 UP000019118 UP000030742 UP000008820 UP000037069 UP000000673 UP000076408 UP000092461 UP000053825 UP000009192 UP000192221 UP000075900 UP000007801 UP000095300 UP000075902 UP000075903 UP000075882 UP000075840 UP000095301 UP000092443 UP000076407 UP000092460 UP000075883 UP000000803 UP000008711 UP000078200 UP000007062 UP000183832
UP000037510 UP000007266 UP000008237 UP000075880 UP000235965 UP000009046 UP000005203 UP000242457 UP000030765 UP000069940 UP000249989 UP000019118 UP000030742 UP000008820 UP000037069 UP000000673 UP000076408 UP000092461 UP000053825 UP000009192 UP000192221 UP000075900 UP000007801 UP000095300 UP000075902 UP000075903 UP000075882 UP000075840 UP000095301 UP000092443 UP000076407 UP000092460 UP000075883 UP000000803 UP000008711 UP000078200 UP000007062 UP000183832
Pfam
PF01663 Phosphodiest
Interpro
SUPFAM
SSF53649
SSF53649
Gene 3D
ProteinModelPortal
H9IX94
A0A3S2TS99
A0A2A4JLJ3
A0A2A4JMI4
A0A194QG85
A0A2H1W6I0
+ More
A0A0N1I8I2 A0A212F9B4 A0A0L7LJR7 A0A139WFE5 E2BQR6 A0A182JE70 A0A2J7PJ72 E0VLL7 A0A088A583 A0A0C9PZY4 A0A1B6J892 A0A1S4F1U4 A0A1B6CPU1 A0A2A3ESJ3 A0A034WNW5 A0A084WKG3 A0A182G324 A0A0A1WGI8 A0A182G332 N6SW48 A0A0K8UNR6 Q17I41 A0A0L0CBV3 W5JUW6 A0A182YKC5 A0A1B0CLM7 A0A0L7QXF1 B4KM92 W8BEH6 A0A1W4U582 A0A182RRX3 A0A1L8DJZ9 B3MIW2 A0A1I8PKM7 A0A182TE79 A0A182UT63 A0A182LLF0 A0A182I126 A0A1I8N2W4 A0A1A9XDF0 A0A182X8Q0 Q7YU86 A0A1B0AMK9 A0A182MCH6 A1ZB84 B3NMT0 A0A1A9UR96 Q7Q5Q3 A0A1J1J8N9 A0A2R7W8K5
A0A0N1I8I2 A0A212F9B4 A0A0L7LJR7 A0A139WFE5 E2BQR6 A0A182JE70 A0A2J7PJ72 E0VLL7 A0A088A583 A0A0C9PZY4 A0A1B6J892 A0A1S4F1U4 A0A1B6CPU1 A0A2A3ESJ3 A0A034WNW5 A0A084WKG3 A0A182G324 A0A0A1WGI8 A0A182G332 N6SW48 A0A0K8UNR6 Q17I41 A0A0L0CBV3 W5JUW6 A0A182YKC5 A0A1B0CLM7 A0A0L7QXF1 B4KM92 W8BEH6 A0A1W4U582 A0A182RRX3 A0A1L8DJZ9 B3MIW2 A0A1I8PKM7 A0A182TE79 A0A182UT63 A0A182LLF0 A0A182I126 A0A1I8N2W4 A0A1A9XDF0 A0A182X8Q0 Q7YU86 A0A1B0AMK9 A0A182MCH6 A1ZB84 B3NMT0 A0A1A9UR96 Q7Q5Q3 A0A1J1J8N9 A0A2R7W8K5
PDB
5UDY
E-value=0.0319166,
Score=91
Ontologies
PATHWAY
GO
PANTHER
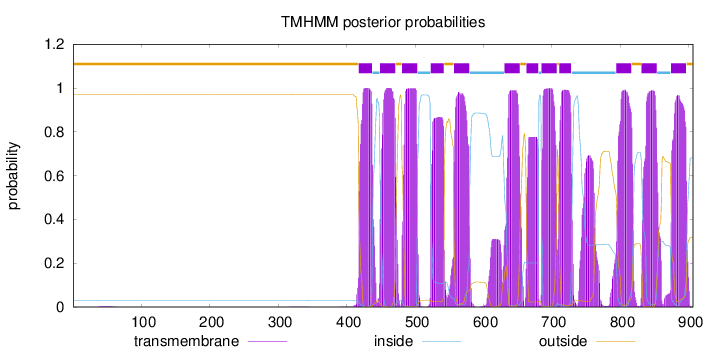
Topology
Length:
906
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
269.3969
Exp number, first 60 AAs:
0.05641
Total prob of N-in:
0.03051
outside
1 - 417
TMhelix
418 - 437
inside
438 - 448
TMhelix
449 - 471
outside
472 - 480
TMhelix
481 - 503
inside
504 - 522
TMhelix
523 - 542
outside
543 - 556
TMhelix
557 - 579
inside
580 - 630
TMhelix
631 - 653
outside
654 - 662
TMhelix
663 - 680
inside
681 - 684
TMhelix
685 - 707
outside
708 - 710
TMhelix
711 - 728
inside
729 - 793
TMhelix
794 - 816
outside
817 - 830
TMhelix
831 - 853
inside
854 - 873
TMhelix
874 - 896
outside
897 - 906
Population Genetic Test Statistics
Pi
218.444099
Theta
175.804034
Tajima's D
0.644308
CLR
0.031973
CSRT
0.565671716414179
Interpretation
Possibly Positive selection
