Gene
KWMTBOMO11407
Pre Gene Modal
BGIBMGA001875
Annotation
putative_lectin_C-type_domain_containing_protein_[Danaus_plexippus]
Full name
Fanconi-associated nuclease
Location in the cell
Nuclear Reliability : 3.049
Sequence
CDS
ATGTTGGGGAGAGCTCATCGAGTGACAGTAACGAATCAGCGAGCTGTTTTAAAAAAAAGAAGTCCAGTCAAAAAGAATTTAGCTCGAGATTCATTTGGATGCTTAGAAAACGATAACTCAATTGATCATGTTTTCAAAGCTGCAAGTGATGGAATGGATGATAAAACTATATTTCTATTGAACATCATCTATAATTTTTTGAATAATGAGAATTTGAAGAACCTACTAGATGAATGTTCTCAGACTCTTTTAAGTAGATGTCTTCATGTGAAAAAGCCTGGTGTCAGAGTTATATGTAGATTGTATTGGAGGCAGGCCACCATTTACCGCTGGGATGATCTTGAAAATATCAGCAATGATAGGTCTAATAAAATTCAAAAACAAATAGTACAAGAAATGATCAGTGATCTAGTGAAAAATGGTTTCCTTGAAAATGTTAATTCTGATGATACATGCAAATTAAATTTTCATGAGATAACTTCTAAATTGAAAGCTGATGAATTGAAACAAATATGCAAGGAATTAAATATAAAGGTTCAGTCCAAACAAAGTGCAATAAAATTATTAGAGAATTACAGCAACCAAAGTTCCATCAGCAAGTTCTTTATACAAGCGAAAGAAAACCCTACCAACAACAAGACCAGACTCTTGGAAATATTTAAAAAAAAATTAGGGTCGTGCTACAAGTTAACTGAAAAAGCAAGAAGTACACTGTATGAGCTGTATCTATTGACATACTTAGGAATGGGCTACTCAATTATAAGAGAGAAGAGGCTGGAACTTACATTACTGTATGAGAAGATTAATCGTGAGACATTTCCAATTATCGAAAACAGGGATATTGATGATGCGAGTGTTGTTTTCAAAAATCGAAAAGAGTTTGAGAAGTATCTAGACGCTCATTATATATACGAAAAGTATATAATCGAGACGGACGATAAAGTCAAATGTATTTTAGTAGAAACGGTGTATGAAATGTACAAGAAAATTGACAAGGAAGACATGATTAGATACACTTCACTACCAGAGTGGTTGCGTCGATATACGCCGCCATATATATTTGTGAAAATATTAGGCGAAGGGATCCAGTCGCTGAAGCGTAATAAAAGTGACGTGTCTTCGAACTTCGCCGTTGAAATACTCAGTGTGCTGATAGCGCAGGACTCGTTCAGGCAACACAAGAAAGCTGGCTGGTATACCGAAAAAGCTTTAATACTACAGAAATATCTTGGACGGCACGATGAAGCGGCCCAGTTATTATTGGAAGGACTTCAGTCCAACAAACTTTCAGACGACGCCAAAGACGCGATGCGACCTCAAGCTGTCAAAATATCGAAGCAGAAAACAAATGTAGGAAACAAACTAAGAATAATTCTCAACGAAATCGCCAATGAAAATATCAAATTGGAAAAGGATTTGAGCGCTGAGCATTTACACAAAAATCCTATGGATAACTGGTATAAAGGCAAATTGAAGTACGAGGTTTGCATAGATGGGATAAGGCAAGCCATTGAGGTTGAAGATTACTGTATCTGGCATTACATTAACAACGGAACGTACACCCACGGTGATCACTGGGAAGGCAGAATAATCACGACAATATTCTTTCTGCTGTTCTGGGATATCATATACTCGAAACCTGCTCACGCACCAGGTATATTCCTCAGTCGCTATCAGAGGTATCCATTGGACTTGTACTGCGATAGTTTCTACACGAATAGAAAGAGTGAGATCGACGAGCGATTGAGTAGTCTAGCGGCGGGCAGCGCAGAGGATATGTTGGAGGCCATGAGACGCGTGTGGGACTCGAGGCCCGAGGGTGAACTATCTGGACTAACGAGGAGCATAGAGTGGGACAGGATAGAGATGGTGGCGGGTTGCCTGGGGCCGCGGCGGGTGGGGGCGCTGTGCGAGCGACTCGCCACGAACTACGGGCACGCGCACAGCGGCTTCCCCGACCTCACGCTGTGGAACGCACACACGAAAACAATAACGTTCATGGAGGTGAAGTCTGACTCAGACCGTCCGTCCGTGAAGCAGCTGCAGTGGATGCACTATCTCCAGCAGCGCGGGCTCCGCACGGCCTTCTGCTACGTCGGCGGAGACTCCGGGCGCCGCGCCAGGCTCGACCGCCCCTGA
Protein
MLGRAHRVTVTNQRAVLKKRSPVKKNLARDSFGCLENDNSIDHVFKAASDGMDDKTIFLLNIIYNFLNNENLKNLLDECSQTLLSRCLHVKKPGVRVICRLYWRQATIYRWDDLENISNDRSNKIQKQIVQEMISDLVKNGFLENVNSDDTCKLNFHEITSKLKADELKQICKELNIKVQSKQSAIKLLENYSNQSSISKFFIQAKENPTNNKTRLLEIFKKKLGSCYKLTEKARSTLYELYLLTYLGMGYSIIREKRLELTLLYEKINRETFPIIENRDIDDASVVFKNRKEFEKYLDAHYIYEKYIIETDDKVKCILVETVYEMYKKIDKEDMIRYTSLPEWLRRYTPPYIFVKILGEGIQSLKRNKSDVSSNFAVEILSVLIAQDSFRQHKKAGWYTEKALILQKYLGRHDEAAQLLLEGLQSNKLSDDAKDAMRPQAVKISKQKTNVGNKLRIILNEIANENIKLEKDLSAEHLHKNPMDNWYKGKLKYEVCIDGIRQAIEVEDYCIWHYINNGTYTHGDHWEGRIITTIFFLLFWDIIYSKPAHAPGIFLSRYQRYPLDLYCDSFYTNRKSEIDERLSSLAAGSAEDMLEAMRRVWDSRPEGELSGLTRSIEWDRIEMVAGCLGPRRVGALCERLATNYGHAHSGFPDLTLWNAHTKTITFMEVKSDSDRPSVKQLQWMHYLQQRGLRTAFCYVGGDSGRRARLDRP
Summary
Description
Nuclease required for the repair of DNA interstrand cross-links (ICL). Acts as a 5'-3' exonuclease that anchors at a cut end of DNA and cleaves DNA successively at every third nucleotide, allowing to excise an ICL from one strand through flanking incisions.
Catalytic Activity
Hydrolytically removes 5'-nucleotides successively from the 3'-hydroxy termini of 3'-hydroxy-terminated oligonucleotides.
Similarity
Belongs to the FAN1 family.
Uniprot
H9IX92
A0A2A4KAH8
A0A212FLR9
A0A1E1W4E8
A0A0N1INX1
A0A194QER5
+ More
A0A2H1WEE6 A0A195EZY0 A0A195BPW9 A0A158P3D6 A0A151IIV5 F4X5G5 K1PPM8 A0A232EQZ2 A0A1B6E1X3 A0A1Y1MXD8 A0A3Q4GYP6 A0A3Q4H5E4 A0A3Q3BKM7 A0A026WUN0 A0A3L8DQN8 V4CF11 A0A3Q0SB63 A0A2J7PE50 A0A067RK85 A0A3P9DS61 A0A151MA71 W4ZGR0 I3JNQ4 A0A3Q2UAY9 A0A3P8UA54 A0A3Q1AVH2 A0A195DNH1 A0A3B4GZH6 A0A3B4ZPU5 A0A151WYM7 A0A3Q3RIZ4 A0A3B4Z8S4 A0A3B4VHW6 A0A1S3JTU7 A0A2H3IJE7 A0A3Q1IPR2 A0A3Q1EZD5 A0A1D1UQM7 A0A3Q3J254 A0A3P8W105 G3NS19 Q4S1C9 A0A1U8CXE0 H2TCT3 A0A3P8XKY7 A0A369SDZ5 K7GHJ6 A0A3P8P8U4 A0A3Q3FIG2 B3RR70 A0A293LFF3 B6QND3 A0A096M710 A0A225AJ22 A0A3B5MCV4 A0A0F8BYR9 A0A364L0F5 A0A3R7IJ89 E9ICP9 A0A315WGK5 A0A397I2X6 A0A3Q3X4D1 M3ZWL0 A0A3B3C8M0 A0A087XG46 H3D860 A0A093UR11 A0A3B3U8R5 A0A2R5LGX3 A0A3B3YTJ3 A0A3P9H6G9 A0A1Q5UHH2 I3JNQ3 A0A1S8BJ35 Q0CIM7 A0A1S3PDI8 A0A3B3HSW7 A0A1Y2ABU1 C5FT18 A0A3Q3BI08 C6HA30
A0A2H1WEE6 A0A195EZY0 A0A195BPW9 A0A158P3D6 A0A151IIV5 F4X5G5 K1PPM8 A0A232EQZ2 A0A1B6E1X3 A0A1Y1MXD8 A0A3Q4GYP6 A0A3Q4H5E4 A0A3Q3BKM7 A0A026WUN0 A0A3L8DQN8 V4CF11 A0A3Q0SB63 A0A2J7PE50 A0A067RK85 A0A3P9DS61 A0A151MA71 W4ZGR0 I3JNQ4 A0A3Q2UAY9 A0A3P8UA54 A0A3Q1AVH2 A0A195DNH1 A0A3B4GZH6 A0A3B4ZPU5 A0A151WYM7 A0A3Q3RIZ4 A0A3B4Z8S4 A0A3B4VHW6 A0A1S3JTU7 A0A2H3IJE7 A0A3Q1IPR2 A0A3Q1EZD5 A0A1D1UQM7 A0A3Q3J254 A0A3P8W105 G3NS19 Q4S1C9 A0A1U8CXE0 H2TCT3 A0A3P8XKY7 A0A369SDZ5 K7GHJ6 A0A3P8P8U4 A0A3Q3FIG2 B3RR70 A0A293LFF3 B6QND3 A0A096M710 A0A225AJ22 A0A3B5MCV4 A0A0F8BYR9 A0A364L0F5 A0A3R7IJ89 E9ICP9 A0A315WGK5 A0A397I2X6 A0A3Q3X4D1 M3ZWL0 A0A3B3C8M0 A0A087XG46 H3D860 A0A093UR11 A0A3B3U8R5 A0A2R5LGX3 A0A3B3YTJ3 A0A3P9H6G9 A0A1Q5UHH2 I3JNQ3 A0A1S8BJ35 Q0CIM7 A0A1S3PDI8 A0A3B3HSW7 A0A1Y2ABU1 C5FT18 A0A3Q3BI08 C6HA30
EC Number
3.1.4.1
Pubmed
19121390
22118469
26354079
21347285
21719571
22992520
+ More
28648823 28004739 25186727 24508170 30249741 23254933 24845553 22293439 28951729 27649274 24487278 15496914 21551351 25069045 30042472 17381049 18719581 25676766 25835551 28649280 21282665 29703783 23542700 29451363 25330172 17554307 22951933
28648823 28004739 25186727 24508170 30249741 23254933 24845553 22293439 28951729 27649274 24487278 15496914 21551351 25069045 30042472 17381049 18719581 25676766 25835551 28649280 21282665 29703783 23542700 29451363 25330172 17554307 22951933
EMBL
BABH01018115
BABH01018116
BABH01018117
NWSH01000016
PCG80793.1
AGBW02007717
+ More
OWR54676.1 GDQN01009245 GDQN01001405 JAT81809.1 JAT89649.1 KQ460772 KPJ12470.1 KQ459053 KPJ03972.1 ODYU01008094 SOQ51397.1 KQ981905 KYN33467.1 KQ976432 KYM87539.1 ADTU01007654 KQ977459 KYN02574.1 GL888716 EGI58321.1 JH815782 EKC26127.1 NNAY01002706 OXU20737.1 GEDC01005365 JAS31933.1 GEZM01024050 JAV88027.1 KK107099 EZA59668.1 QOIP01000005 RLU22755.1 KB200701 ESP00585.1 NEVH01026109 PNF14614.1 KK852461 KDR23438.1 AKHW03006295 KYO21391.1 AAGJ04163082 AERX01010884 KQ980713 KYN14377.1 KQ982649 KYQ52988.1 NPFK01000054 PCH04433.1 BDGG01000002 GAU91751.1 CAAE01014769 CAG05553.1 NOWV01000018 RDD45148.1 AGCU01176510 AGCU01176511 AGCU01176512 DS985243 EDV26826.1 GFWV01002752 MAA27482.1 DS995903 EEA21421.1 AYCK01018417 LFMY01000007 OKL59343.1 KQ041939 KKF20760.1 MIKG01000009 RAO69293.1 NIDN02000090 RLL97047.1 GL762354 EFZ21646.1 NHOQ01000384 PWA30476.1 NKHV02000033 RHZ69895.1 JPOX01000045 KFX42365.1 GGLE01004640 MBY08766.1 MNBE01000259 OKP11903.1 MSZU01000076 OMP87486.1 CH476602 EAU33001.1 MCFA01000002 ORY19485.1 DS995705 EEQ33021.1 GG692421 EER43163.1
OWR54676.1 GDQN01009245 GDQN01001405 JAT81809.1 JAT89649.1 KQ460772 KPJ12470.1 KQ459053 KPJ03972.1 ODYU01008094 SOQ51397.1 KQ981905 KYN33467.1 KQ976432 KYM87539.1 ADTU01007654 KQ977459 KYN02574.1 GL888716 EGI58321.1 JH815782 EKC26127.1 NNAY01002706 OXU20737.1 GEDC01005365 JAS31933.1 GEZM01024050 JAV88027.1 KK107099 EZA59668.1 QOIP01000005 RLU22755.1 KB200701 ESP00585.1 NEVH01026109 PNF14614.1 KK852461 KDR23438.1 AKHW03006295 KYO21391.1 AAGJ04163082 AERX01010884 KQ980713 KYN14377.1 KQ982649 KYQ52988.1 NPFK01000054 PCH04433.1 BDGG01000002 GAU91751.1 CAAE01014769 CAG05553.1 NOWV01000018 RDD45148.1 AGCU01176510 AGCU01176511 AGCU01176512 DS985243 EDV26826.1 GFWV01002752 MAA27482.1 DS995903 EEA21421.1 AYCK01018417 LFMY01000007 OKL59343.1 KQ041939 KKF20760.1 MIKG01000009 RAO69293.1 NIDN02000090 RLL97047.1 GL762354 EFZ21646.1 NHOQ01000384 PWA30476.1 NKHV02000033 RHZ69895.1 JPOX01000045 KFX42365.1 GGLE01004640 MBY08766.1 MNBE01000259 OKP11903.1 MSZU01000076 OMP87486.1 CH476602 EAU33001.1 MCFA01000002 ORY19485.1 DS995705 EEQ33021.1 GG692421 EER43163.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000078541
+ More
UP000078540 UP000005205 UP000078542 UP000007755 UP000005408 UP000215335 UP000261580 UP000264840 UP000053097 UP000279307 UP000030746 UP000261340 UP000235965 UP000027135 UP000265160 UP000050525 UP000007110 UP000005207 UP000265000 UP000265080 UP000257160 UP000078492 UP000261460 UP000261400 UP000075809 UP000261640 UP000261420 UP000085678 UP000218381 UP000265040 UP000257200 UP000186922 UP000261600 UP000265120 UP000007635 UP000189705 UP000005226 UP000265140 UP000253843 UP000007267 UP000265100 UP000261660 UP000009022 UP000001294 UP000028760 UP000214365 UP000261380 UP000249363 UP000215289 UP000215394 UP000261620 UP000002852 UP000261560 UP000007303 UP000029285 UP000261500 UP000261480 UP000265200 UP000186955 UP000190776 UP000007963 UP000087266 UP000001038 UP000193144 UP000002035 UP000264800 UP000002624
UP000078540 UP000005205 UP000078542 UP000007755 UP000005408 UP000215335 UP000261580 UP000264840 UP000053097 UP000279307 UP000030746 UP000261340 UP000235965 UP000027135 UP000265160 UP000050525 UP000007110 UP000005207 UP000265000 UP000265080 UP000257160 UP000078492 UP000261460 UP000261400 UP000075809 UP000261640 UP000261420 UP000085678 UP000218381 UP000265040 UP000257200 UP000186922 UP000261600 UP000265120 UP000007635 UP000189705 UP000005226 UP000265140 UP000253843 UP000007267 UP000265100 UP000261660 UP000009022 UP000001294 UP000028760 UP000214365 UP000261380 UP000249363 UP000215289 UP000215394 UP000261620 UP000002852 UP000261560 UP000007303 UP000029285 UP000261500 UP000261480 UP000265200 UP000186955 UP000190776 UP000007963 UP000087266 UP000001038 UP000193144 UP000002035 UP000264800 UP000002624
PRIDE
Pfam
PF08774 VRR_NUC
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9IX92
A0A2A4KAH8
A0A212FLR9
A0A1E1W4E8
A0A0N1INX1
A0A194QER5
+ More
A0A2H1WEE6 A0A195EZY0 A0A195BPW9 A0A158P3D6 A0A151IIV5 F4X5G5 K1PPM8 A0A232EQZ2 A0A1B6E1X3 A0A1Y1MXD8 A0A3Q4GYP6 A0A3Q4H5E4 A0A3Q3BKM7 A0A026WUN0 A0A3L8DQN8 V4CF11 A0A3Q0SB63 A0A2J7PE50 A0A067RK85 A0A3P9DS61 A0A151MA71 W4ZGR0 I3JNQ4 A0A3Q2UAY9 A0A3P8UA54 A0A3Q1AVH2 A0A195DNH1 A0A3B4GZH6 A0A3B4ZPU5 A0A151WYM7 A0A3Q3RIZ4 A0A3B4Z8S4 A0A3B4VHW6 A0A1S3JTU7 A0A2H3IJE7 A0A3Q1IPR2 A0A3Q1EZD5 A0A1D1UQM7 A0A3Q3J254 A0A3P8W105 G3NS19 Q4S1C9 A0A1U8CXE0 H2TCT3 A0A3P8XKY7 A0A369SDZ5 K7GHJ6 A0A3P8P8U4 A0A3Q3FIG2 B3RR70 A0A293LFF3 B6QND3 A0A096M710 A0A225AJ22 A0A3B5MCV4 A0A0F8BYR9 A0A364L0F5 A0A3R7IJ89 E9ICP9 A0A315WGK5 A0A397I2X6 A0A3Q3X4D1 M3ZWL0 A0A3B3C8M0 A0A087XG46 H3D860 A0A093UR11 A0A3B3U8R5 A0A2R5LGX3 A0A3B3YTJ3 A0A3P9H6G9 A0A1Q5UHH2 I3JNQ3 A0A1S8BJ35 Q0CIM7 A0A1S3PDI8 A0A3B3HSW7 A0A1Y2ABU1 C5FT18 A0A3Q3BI08 C6HA30
A0A2H1WEE6 A0A195EZY0 A0A195BPW9 A0A158P3D6 A0A151IIV5 F4X5G5 K1PPM8 A0A232EQZ2 A0A1B6E1X3 A0A1Y1MXD8 A0A3Q4GYP6 A0A3Q4H5E4 A0A3Q3BKM7 A0A026WUN0 A0A3L8DQN8 V4CF11 A0A3Q0SB63 A0A2J7PE50 A0A067RK85 A0A3P9DS61 A0A151MA71 W4ZGR0 I3JNQ4 A0A3Q2UAY9 A0A3P8UA54 A0A3Q1AVH2 A0A195DNH1 A0A3B4GZH6 A0A3B4ZPU5 A0A151WYM7 A0A3Q3RIZ4 A0A3B4Z8S4 A0A3B4VHW6 A0A1S3JTU7 A0A2H3IJE7 A0A3Q1IPR2 A0A3Q1EZD5 A0A1D1UQM7 A0A3Q3J254 A0A3P8W105 G3NS19 Q4S1C9 A0A1U8CXE0 H2TCT3 A0A3P8XKY7 A0A369SDZ5 K7GHJ6 A0A3P8P8U4 A0A3Q3FIG2 B3RR70 A0A293LFF3 B6QND3 A0A096M710 A0A225AJ22 A0A3B5MCV4 A0A0F8BYR9 A0A364L0F5 A0A3R7IJ89 E9ICP9 A0A315WGK5 A0A397I2X6 A0A3Q3X4D1 M3ZWL0 A0A3B3C8M0 A0A087XG46 H3D860 A0A093UR11 A0A3B3U8R5 A0A2R5LGX3 A0A3B3YTJ3 A0A3P9H6G9 A0A1Q5UHH2 I3JNQ3 A0A1S8BJ35 Q0CIM7 A0A1S3PDI8 A0A3B3HSW7 A0A1Y2ABU1 C5FT18 A0A3Q3BI08 C6HA30
PDB
4RIC
E-value=1.51057e-31,
Score=343
Ontologies
GO
GO:0004528
GO:0003676
GO:0036297
GO:0005634
GO:0046872
GO:0006281
GO:0017108
GO:0008409
GO:0070336
GO:0003677
GO:0000724
GO:0033683
GO:0045171
GO:0005654
GO:0005829
GO:0140036
GO:0016021
GO:0016788
GO:0016491
GO:0006313
GO:0007018
GO:0030286
GO:0046034
GO:0032559
GO:0004812
GO:0006418
GO:0003824
GO:0004518
PANTHER
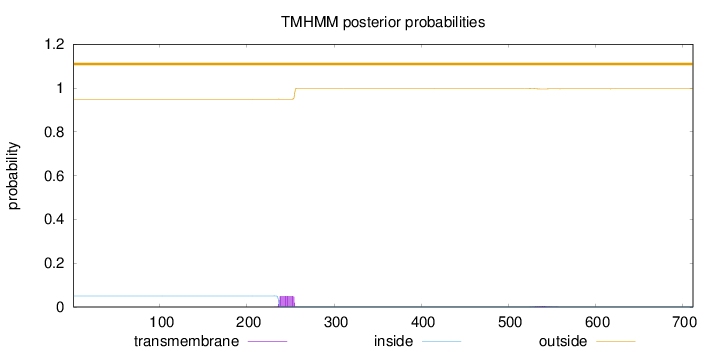
Topology
Subcellular location
Nucleus
Length:
712
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.996589999999999
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.05067
outside
1 - 712
Population Genetic Test Statistics
Pi
230.106268
Theta
175.028345
Tajima's D
0
CLR
0.618895
CSRT
0.369531523423829
Interpretation
Possibly Positive selection
