Gene
KWMTBOMO11405 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001874
Annotation
vesicle_amine_transport_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.848 Nuclear Reliability : 1.189
Sequence
CDS
ATGACTGAGAGTACGGACGCCGCCGCGACTCCAGCACCGGAGAAGACCCCGACCGAAAATGGTGCTACTGAAACACCGCAAGAGGAAACGCCCGAAAAATCTACCGAGGAGAAAAAGGATGAGACCCCACCGCCCAAGGAAATGCGCGCCGTAGTACTTACCGGCTTCGGGGGTCTCAAGACCGTTAAGATACTCAAAAAACCTGAGCCGACCGTTGGTGAGGGCGAGGTCCTGATTCGCGTGAAAGCCTGCGGCCTAAACTTCCAAGATTTGATAGTTCGTCAGGGCGCCATCGACTCTCCACCGAAGACTCCTTTCATCTTGGGCTTCGAATGCGCTGGTGAAATCGAGCAAGTTGGCGAAAATGTCACCAATTTTAAGGTGGGCGATCAAGTGGTGGCTCTTCCCGAGTACCGCGCTTGGGCCGAGCTGGTATCTGTACCGGCGCAGTATGTGTACGCGCTGCCCGAAGGAATGTCTGCCCTGGATGCTGTTGCCATCACCACGAACTACGTGGTGGCCTATCTGCTGCTCTTCGAGATGGCTAACTTGACTCCCGGCAAGAGTCTACTCGTGCATTCCGCTGGAGGCGGTGTGGGACAAGCTGTAGCCCAGTTGGCGAAGACGGTCGAAAACGTGACCGTGTTTGGTGTCTGCTCCAAAAGCAAGCACGAGGCGCTGAAGACAAACAACAATAACATCGACCACCTGCTCGAGAGGGGCAGCGACTACACCAGCGAAGTCAGGAAGGTAACTCCGGATGGCGTAGACATTGTGCTGGATTGCCTCTGCGGCGAGGAGTGCAACCGCGGGTATTCGCTTCTGAAGCCAATGGGCCGTTACATCCTCTATGGATCATCTAACATCGTGACCGGCGAGACAAAGAGCTTCTTCAGCGCTGCCCGTGCGTGGTGGCAAGTGGACAAGGTGTCTCCGATCAAGCTGTTCGACGAGAACAAGAGCCTGGCGGGACTGAACCTGCGACACCTGCTGTTCCAGCACGGGCGCGGGGACACGGTGCGGCGCGCCGTGCACAGCGTCTTCACTCTGTGGGCGCAGGGCAAGGTCAAGCCCATCGTCGACTCCACGTGGGCCCTCGAGGATGTGGGCGAAGCCATGCAGAAGATGCACGACCGCAAGAACATCGGCAAGCTCGTCCTGGACCCGTCCCTCGAGCCGAAGCCTAAGCCGGCCACTCCCGCTAAAGGCAAATCCGGCAAGGAGAAGAAGCCGGCCAAGGAGAGCTCTGAGGAGAAGAAGGACAAGGAGGGCAAGGACGATGAGAAAAAGCCAGCCGAAAACGGGGACAAGAACGAGTCGAAGGAGAAGGAGAAAGAGTCTAGCTGA
Protein
MTESTDAAATPAPEKTPTENGATETPQEETPEKSTEEKKDETPPPKEMRAVVLTGFGGLKTVKILKKPEPTVGEGEVLIRVKACGLNFQDLIVRQGAIDSPPKTPFILGFECAGEIEQVGENVTNFKVGDQVVALPEYRAWAELVSVPAQYVYALPEGMSALDAVAITTNYVVAYLLLFEMANLTPGKSLLVHSAGGGVGQAVAQLAKTVENVTVFGVCSKSKHEALKTNNNNIDHLLERGSDYTSEVRKVTPDGVDIVLDCLCGEECNRGYSLLKPMGRYILYGSSNIVTGETKSFFSAARAWWQVDKVSPIKLFDENKSLAGLNLRHLLFQHGRGDTVRRAVHSVFTLWAQGKVKPIVDSTWALEDVGEAMQKMHDRKNIGKLVLDPSLEPKPKPATPAKGKSGKEKKPAKESSEEKKDKEGKDDEKKPAENGDKNESKEKEKESS
Summary
Uniprot
Q1HPK0
S4PM64
A0A2A4K9C1
A0A2A4K9Z9
A0A3S2PL40
A0A0J7L1B8
+ More
A0A088A347 V9IAR8 A0A3L8DRV2 V5GJ73 A0A139WFI3 D6WUA2 A0A151XC74 A0A1Y1NAW0 A0A1L8E0P3 A0A1L8E163 A0A084WMH5 A0A2M4ARY5 Q7Q5E7 A0A2M4CJN3 T1E9F3 A0A2M4CT77 A0A2S2N727 U5EL92 J9JNB2 E0VGB3 A0A1Q3FUI5 A0A1Q3FUL3 A0A1Q3FUR2 A0A1B6M2Q2 A0A2A3E8V4 A0A336KDY2 A0A0P6IJL0 A0A1Z5L730 A0A0P6FEQ6 A0A0P6G4K7 A0A0P6H0U9 A0A1Z5L7E0 L7LZT0 A0A2R5LFB5 L7M5D0 A0A131YDT6 A0A224Z7I1 A0A131YS22 A0A131XLP1 A0A023GBN3 A0A131XZT3 A0A147B873 A0A023FV99 A0A0K8RCN0 A0A1E1XE71 A0A131XTI5 A0A023FLE5 A0A0A9VUS7 A0A154PG01 A0A1S3IJC0 T1IYY4 A0A2P6L4K2 A0A210PE10 A0A2C9KKV4 A0A0B6ZIY0 T1EER8 A0A0L8GM60 A0A0L8GLU8 A0A0L8GMT0 A0A0L8GLW7
A0A088A347 V9IAR8 A0A3L8DRV2 V5GJ73 A0A139WFI3 D6WUA2 A0A151XC74 A0A1Y1NAW0 A0A1L8E0P3 A0A1L8E163 A0A084WMH5 A0A2M4ARY5 Q7Q5E7 A0A2M4CJN3 T1E9F3 A0A2M4CT77 A0A2S2N727 U5EL92 J9JNB2 E0VGB3 A0A1Q3FUI5 A0A1Q3FUL3 A0A1Q3FUR2 A0A1B6M2Q2 A0A2A3E8V4 A0A336KDY2 A0A0P6IJL0 A0A1Z5L730 A0A0P6FEQ6 A0A0P6G4K7 A0A0P6H0U9 A0A1Z5L7E0 L7LZT0 A0A2R5LFB5 L7M5D0 A0A131YDT6 A0A224Z7I1 A0A131YS22 A0A131XLP1 A0A023GBN3 A0A131XZT3 A0A147B873 A0A023FV99 A0A0K8RCN0 A0A1E1XE71 A0A131XTI5 A0A023FLE5 A0A0A9VUS7 A0A154PG01 A0A1S3IJC0 T1IYY4 A0A2P6L4K2 A0A210PE10 A0A2C9KKV4 A0A0B6ZIY0 T1EER8 A0A0L8GM60 A0A0L8GLU8 A0A0L8GMT0 A0A0L8GLW7
Pubmed
EMBL
DQ443402
ABF51491.1
GAIX01003725
JAA88835.1
NWSH01000016
PCG80807.1
+ More
PCG80806.1 RSAL01000004 RVE54533.1 LBMM01001402 KMQ96383.1 JR038245 JR038246 AEY58175.1 AEY58176.1 QOIP01000005 RLU23032.1 GALX01004342 JAB64124.1 KQ971352 KYB26740.1 EFA06248.1 KQ982314 KYQ57981.1 GEZM01007618 JAV95044.1 GFDF01001771 JAV12313.1 GFDF01001772 JAV12312.1 ATLV01024454 ATLV01024455 ATLV01024456 ATLV01024457 ATLV01024458 KE525352 KFB51419.1 GGFK01010245 MBW43566.1 AAAB01008960 EAA11101.4 GGFL01001359 MBW65537.1 GAMD01001412 JAB00179.1 GGFL01003860 MBW68038.1 GGMR01000404 MBY13023.1 GANO01001506 JAB58365.1 ABLF02037614 DS235135 EEB12419.1 GFDL01003818 JAV31227.1 GFDL01003817 JAV31228.1 GFDL01003819 JAV31226.1 GEBQ01009799 JAT30178.1 KZ288322 PBC28155.1 UFQS01000244 UFQT01000244 SSX01948.1 SSX22325.1 GDIQ01003371 JAN91366.1 GFJQ02003764 JAW03206.1 GDIQ01048741 JAN45996.1 GDIQ01050202 JAN44535.1 GDIQ01047243 JAN47494.1 GFJQ02003765 JAW03205.1 GACK01008631 JAA56403.1 GGLE01003989 MBY08115.1 GACK01006730 JAA58304.1 GEDV01011073 JAP77484.1 GFPF01012195 MAA23341.1 GEDV01007179 JAP81378.1 GEFH01000582 JAP67999.1 GBBM01003797 JAC31621.1 GEFM01004665 JAP71131.1 GEIB01001205 JAR86943.1 GBBL01002657 JAC24663.1 GADI01004972 JAA68836.1 GFAC01001767 JAT97421.1 GEFM01005173 JAP70623.1 GBBK01002632 JAC21850.1 GBHO01044145 GBHO01044143 JAF99458.1 JAF99460.1 KQ434896 KZC10793.1 JH431701 MWRG01001856 PRD33505.1 NEDP02076750 OWF34730.1 HACG01021578 CEK68443.1 AMQM01002786 KB095858 ESO10962.1 KQ421261 KOF77944.1 KOF77942.1 KOF77945.1 KOF77943.1
PCG80806.1 RSAL01000004 RVE54533.1 LBMM01001402 KMQ96383.1 JR038245 JR038246 AEY58175.1 AEY58176.1 QOIP01000005 RLU23032.1 GALX01004342 JAB64124.1 KQ971352 KYB26740.1 EFA06248.1 KQ982314 KYQ57981.1 GEZM01007618 JAV95044.1 GFDF01001771 JAV12313.1 GFDF01001772 JAV12312.1 ATLV01024454 ATLV01024455 ATLV01024456 ATLV01024457 ATLV01024458 KE525352 KFB51419.1 GGFK01010245 MBW43566.1 AAAB01008960 EAA11101.4 GGFL01001359 MBW65537.1 GAMD01001412 JAB00179.1 GGFL01003860 MBW68038.1 GGMR01000404 MBY13023.1 GANO01001506 JAB58365.1 ABLF02037614 DS235135 EEB12419.1 GFDL01003818 JAV31227.1 GFDL01003817 JAV31228.1 GFDL01003819 JAV31226.1 GEBQ01009799 JAT30178.1 KZ288322 PBC28155.1 UFQS01000244 UFQT01000244 SSX01948.1 SSX22325.1 GDIQ01003371 JAN91366.1 GFJQ02003764 JAW03206.1 GDIQ01048741 JAN45996.1 GDIQ01050202 JAN44535.1 GDIQ01047243 JAN47494.1 GFJQ02003765 JAW03205.1 GACK01008631 JAA56403.1 GGLE01003989 MBY08115.1 GACK01006730 JAA58304.1 GEDV01011073 JAP77484.1 GFPF01012195 MAA23341.1 GEDV01007179 JAP81378.1 GEFH01000582 JAP67999.1 GBBM01003797 JAC31621.1 GEFM01004665 JAP71131.1 GEIB01001205 JAR86943.1 GBBL01002657 JAC24663.1 GADI01004972 JAA68836.1 GFAC01001767 JAT97421.1 GEFM01005173 JAP70623.1 GBBK01002632 JAC21850.1 GBHO01044145 GBHO01044143 JAF99458.1 JAF99460.1 KQ434896 KZC10793.1 JH431701 MWRG01001856 PRD33505.1 NEDP02076750 OWF34730.1 HACG01021578 CEK68443.1 AMQM01002786 KB095858 ESO10962.1 KQ421261 KOF77944.1 KOF77942.1 KOF77945.1 KOF77943.1
Proteomes
PRIDE
Pfam
PF08240 ADH_N
Interpro
ProteinModelPortal
Q1HPK0
S4PM64
A0A2A4K9C1
A0A2A4K9Z9
A0A3S2PL40
A0A0J7L1B8
+ More
A0A088A347 V9IAR8 A0A3L8DRV2 V5GJ73 A0A139WFI3 D6WUA2 A0A151XC74 A0A1Y1NAW0 A0A1L8E0P3 A0A1L8E163 A0A084WMH5 A0A2M4ARY5 Q7Q5E7 A0A2M4CJN3 T1E9F3 A0A2M4CT77 A0A2S2N727 U5EL92 J9JNB2 E0VGB3 A0A1Q3FUI5 A0A1Q3FUL3 A0A1Q3FUR2 A0A1B6M2Q2 A0A2A3E8V4 A0A336KDY2 A0A0P6IJL0 A0A1Z5L730 A0A0P6FEQ6 A0A0P6G4K7 A0A0P6H0U9 A0A1Z5L7E0 L7LZT0 A0A2R5LFB5 L7M5D0 A0A131YDT6 A0A224Z7I1 A0A131YS22 A0A131XLP1 A0A023GBN3 A0A131XZT3 A0A147B873 A0A023FV99 A0A0K8RCN0 A0A1E1XE71 A0A131XTI5 A0A023FLE5 A0A0A9VUS7 A0A154PG01 A0A1S3IJC0 T1IYY4 A0A2P6L4K2 A0A210PE10 A0A2C9KKV4 A0A0B6ZIY0 T1EER8 A0A0L8GM60 A0A0L8GLU8 A0A0L8GMT0 A0A0L8GLW7
A0A088A347 V9IAR8 A0A3L8DRV2 V5GJ73 A0A139WFI3 D6WUA2 A0A151XC74 A0A1Y1NAW0 A0A1L8E0P3 A0A1L8E163 A0A084WMH5 A0A2M4ARY5 Q7Q5E7 A0A2M4CJN3 T1E9F3 A0A2M4CT77 A0A2S2N727 U5EL92 J9JNB2 E0VGB3 A0A1Q3FUI5 A0A1Q3FUL3 A0A1Q3FUR2 A0A1B6M2Q2 A0A2A3E8V4 A0A336KDY2 A0A0P6IJL0 A0A1Z5L730 A0A0P6FEQ6 A0A0P6G4K7 A0A0P6H0U9 A0A1Z5L7E0 L7LZT0 A0A2R5LFB5 L7M5D0 A0A131YDT6 A0A224Z7I1 A0A131YS22 A0A131XLP1 A0A023GBN3 A0A131XZT3 A0A147B873 A0A023FV99 A0A0K8RCN0 A0A1E1XE71 A0A131XTI5 A0A023FLE5 A0A0A9VUS7 A0A154PG01 A0A1S3IJC0 T1IYY4 A0A2P6L4K2 A0A210PE10 A0A2C9KKV4 A0A0B6ZIY0 T1EER8 A0A0L8GM60 A0A0L8GLU8 A0A0L8GMT0 A0A0L8GLW7
PDB
4A27
E-value=2.05897e-112,
Score=1038
Ontologies
GO
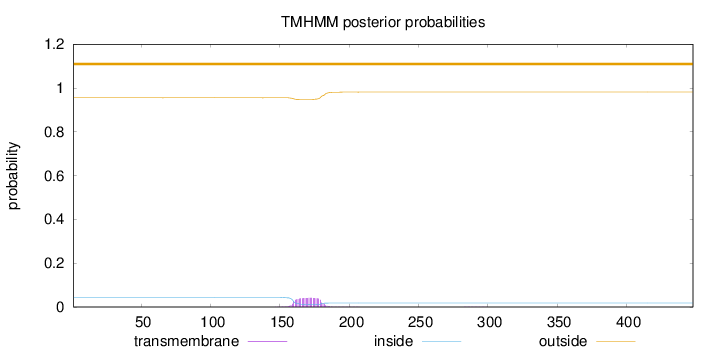
Topology
Length:
448
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.89821
Exp number, first 60 AAs:
0.00026
Total prob of N-in:
0.04469
outside
1 - 448
Population Genetic Test Statistics
Pi
199.58444
Theta
157.507511
Tajima's D
0.757014
CLR
0.448501
CSRT
0.584670766461677
Interpretation
Uncertain
