Gene
KWMTBOMO11404
Pre Gene Modal
BGIBMGA001872
Annotation
PREDICTED:_putative_ribosome-binding_factor_A?_mitochondrial_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.801
Sequence
CDS
ATGGTTAATCCGAAACCAAAAAAGCACTGGTATGCACCTTCAATGTTAGAATTCAACAAAATGCCCTCTGTGAAATCTTTAACAAAAGTTACCCATGAACCCGGCAAGCGTGGAATACGCCGAGTGGCGATGCTGAACAAGATGTTTATGAAACACATTACTGATCTGATGTCCACAGGAACGGTTTCAATGGATATTATCGGACGAGGGATTGAAATATCAAAAGTGAATGTAACATCTGATTTTCAAACTGTACATGTTTATTGGATATGCAAAGGTACCTCGACAGATGAAGAGACGGAAGCCACATTGAACTCTGTTGCAGGAGCTCTGAGGCATGAGCTGTCTGTGCTCAGGATCATGGGCCAGGTTCCTTATATTGTCTTTGTTAAAGATATGCATGAAGCCCGGATATTAGATCTCGATAATAGATTGCTCAAAGCTGATTTTGGAGAAAACTACACACCAACTGATCCAGGGCATTTACTTAAGACAGAATTCACACTAGACACCAAAATATCTCCTGACATCAAGGCCAAAATACGGCAGTTAGAAATAGACGAACCAAGTCAAGAGAGTCCAATACCAGAAATGACTCACACGATCTACGGCTTAGACCACGCCAAGATAATGAACAGACTCTTAACTGCTAGGAAGAAAAGTAAAGACGCTTGGAGTAATCTTGAATCAGAATCGCCAGTTATATCTTACAGAATACATGAAAGCACACCTATCAAAGTCGATACTGTCACCCAAAATAAGGAATTGGCTGAGTTTTTACTTAAAAGACAGATACTACAAAATAAATTGGCCAAGCAGTTACGGAAAACAAGCGACGACTGGCAGTTGTCGGACGATAGCGGATCCAATTCCGACGAGGAGTACATTGATGATACCTACTACGAAGACGAGGAGTATTATTACGATGATATAGAAGAAAATAAGAAAAAATTTGCCAAAGAATAA
Protein
MVNPKPKKHWYAPSMLEFNKMPSVKSLTKVTHEPGKRGIRRVAMLNKMFMKHITDLMSTGTVSMDIIGRGIEISKVNVTSDFQTVHVYWICKGTSTDEETEATLNSVAGALRHELSVLRIMGQVPYIVFVKDMHEARILDLDNRLLKADFGENYTPTDPGHLLKTEFTLDTKISPDIKAKIRQLEIDEPSQESPIPEMTHTIYGLDHAKIMNRLLTARKKSKDAWSNLESESPVISYRIHESTPIKVDTVTQNKELAEFLLKRQILQNKLAKQLRKTSDDWQLSDDSGSNSDEEYIDDTYYEDEEYYYDDIEENKKKFAKE
Summary
Uniprot
H9IX89
A0A194QKJ2
A0A0N1I8H5
A0A2A4K9Q5
A0A2H1W1J9
A0A212FLR8
+ More
A0A2J7PJ95 A0A2J7PJ79 J9JY97 A0A1J1IIC7 A0A2H8TSM7 A0A182GP75 D6WR11 B0WY21 A0A1B0CXC2 Q16IL9 A0A2S2PV88 A0A1W4WYI0 A0A1B0EZK3 U4UEQ8 N6U904 K7J115 A0A1Y1JX99 A0A0K8TSD0 A0A1B6C1M9 A0A3Q0IIN9 A0A182IY11 A0A1I8MY57 B4NEP5 A0A0M4EYA8 A0A182RNQ2 T1PAQ8 A0A0C9REP4 A0A1I8Q5S1 A0A1A9VSZ3 A0A232F5K6 A0A1B0G9T9 A0A084VRD0 A0A1Y1JU48 A0A1B6KVE5 A0A0L0C524 B4L8U1 A0A2L2Y635 A0A1A9YM76 A0A1B6FTU9 A0A034WGL4 A0A1B0A1V0 D3TR07 A0A0K8U891 A0A1B0BII4 A0A1A9W1Z3 W8C0D0 Q8SXX0 B4PX89 B3NXH9 A0A0L7QQH4 E1ZYV8 A0A310SIF7 A0A151XCA9 B4JNV4 B3M5F7 A0A195F2B4 A0A158NFT7
A0A2J7PJ95 A0A2J7PJ79 J9JY97 A0A1J1IIC7 A0A2H8TSM7 A0A182GP75 D6WR11 B0WY21 A0A1B0CXC2 Q16IL9 A0A2S2PV88 A0A1W4WYI0 A0A1B0EZK3 U4UEQ8 N6U904 K7J115 A0A1Y1JX99 A0A0K8TSD0 A0A1B6C1M9 A0A3Q0IIN9 A0A182IY11 A0A1I8MY57 B4NEP5 A0A0M4EYA8 A0A182RNQ2 T1PAQ8 A0A0C9REP4 A0A1I8Q5S1 A0A1A9VSZ3 A0A232F5K6 A0A1B0G9T9 A0A084VRD0 A0A1Y1JU48 A0A1B6KVE5 A0A0L0C524 B4L8U1 A0A2L2Y635 A0A1A9YM76 A0A1B6FTU9 A0A034WGL4 A0A1B0A1V0 D3TR07 A0A0K8U891 A0A1B0BII4 A0A1A9W1Z3 W8C0D0 Q8SXX0 B4PX89 B3NXH9 A0A0L7QQH4 E1ZYV8 A0A310SIF7 A0A151XCA9 B4JNV4 B3M5F7 A0A195F2B4 A0A158NFT7
Pubmed
19121390
26354079
22118469
26483478
18362917
19820115
+ More
17510324 23537049 20075255 28004739 26369729 25315136 17994087 28648823 24438588 26108605 26561354 25348373 20353571 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20798317 21347285
17510324 23537049 20075255 28004739 26369729 25315136 17994087 28648823 24438588 26108605 26561354 25348373 20353571 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20798317 21347285
EMBL
BABH01018123
KQ459053
KPJ03976.1
KQ460772
KPJ12465.1
NWSH01000016
+ More
PCG80809.1 ODYU01005757 SOQ46938.1 AGBW02007717 OWR54682.1 NEVH01024950 PNF16398.1 PNF16397.1 ABLF02020222 CVRI01000047 CRK98209.1 GFXV01005429 MBW17234.1 JXUM01077850 KQ563030 KXJ74613.1 KQ971351 EFA06539.1 DS232179 EDS36831.1 AJWK01033526 CH478071 EAT34112.1 GGMS01000100 MBY69303.1 AJVK01017265 AJVK01017266 AJVK01017267 KB632326 ERL92454.1 APGK01035089 KB740923 ENN78170.1 GEZM01100620 JAV52841.1 GDAI01000557 JAI17046.1 GEDC01029900 JAS07398.1 CH964239 EDW82214.1 CP012528 ALC48942.1 KA644998 AFP59627.1 GBYB01015074 JAG84841.1 NNAY01000922 OXU25892.1 CCAG010011658 ATLV01015606 KE525023 KFB40524.1 GEZM01100622 JAV52839.1 GEBQ01024582 JAT15395.1 JRES01000981 KNC26539.1 CH933815 EDW08066.1 IAAA01017217 IAAA01017218 LAA03624.1 GECZ01016113 JAS53656.1 GAKP01005113 JAC53839.1 EZ423859 ADD20135.1 GDHF01029739 JAI22575.1 JXJN01014969 GAMC01004082 JAC02474.1 AE014298 AY075537 AAF48535.2 AAL68344.1 CM000162 EDX01852.2 CH954180 EDV46938.2 KQ414786 KOC60868.1 GL435242 EFN73648.1 KQ765605 OAD53859.1 KQ982314 KYQ57979.1 CH916371 EDV92397.1 CH902618 EDV39567.2 KQ981864 KYN34317.1 ADTU01014727
PCG80809.1 ODYU01005757 SOQ46938.1 AGBW02007717 OWR54682.1 NEVH01024950 PNF16398.1 PNF16397.1 ABLF02020222 CVRI01000047 CRK98209.1 GFXV01005429 MBW17234.1 JXUM01077850 KQ563030 KXJ74613.1 KQ971351 EFA06539.1 DS232179 EDS36831.1 AJWK01033526 CH478071 EAT34112.1 GGMS01000100 MBY69303.1 AJVK01017265 AJVK01017266 AJVK01017267 KB632326 ERL92454.1 APGK01035089 KB740923 ENN78170.1 GEZM01100620 JAV52841.1 GDAI01000557 JAI17046.1 GEDC01029900 JAS07398.1 CH964239 EDW82214.1 CP012528 ALC48942.1 KA644998 AFP59627.1 GBYB01015074 JAG84841.1 NNAY01000922 OXU25892.1 CCAG010011658 ATLV01015606 KE525023 KFB40524.1 GEZM01100622 JAV52839.1 GEBQ01024582 JAT15395.1 JRES01000981 KNC26539.1 CH933815 EDW08066.1 IAAA01017217 IAAA01017218 LAA03624.1 GECZ01016113 JAS53656.1 GAKP01005113 JAC53839.1 EZ423859 ADD20135.1 GDHF01029739 JAI22575.1 JXJN01014969 GAMC01004082 JAC02474.1 AE014298 AY075537 AAF48535.2 AAL68344.1 CM000162 EDX01852.2 CH954180 EDV46938.2 KQ414786 KOC60868.1 GL435242 EFN73648.1 KQ765605 OAD53859.1 KQ982314 KYQ57979.1 CH916371 EDV92397.1 CH902618 EDV39567.2 KQ981864 KYN34317.1 ADTU01014727
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000235965
+ More
UP000007819 UP000183832 UP000069940 UP000249989 UP000007266 UP000002320 UP000092461 UP000008820 UP000192223 UP000092462 UP000030742 UP000019118 UP000002358 UP000079169 UP000075880 UP000095301 UP000007798 UP000092553 UP000075900 UP000095300 UP000078200 UP000215335 UP000092444 UP000030765 UP000037069 UP000009192 UP000092443 UP000092445 UP000092460 UP000091820 UP000000803 UP000002282 UP000008711 UP000053825 UP000000311 UP000075809 UP000001070 UP000007801 UP000078541 UP000005205
UP000007819 UP000183832 UP000069940 UP000249989 UP000007266 UP000002320 UP000092461 UP000008820 UP000192223 UP000092462 UP000030742 UP000019118 UP000002358 UP000079169 UP000075880 UP000095301 UP000007798 UP000092553 UP000075900 UP000095300 UP000078200 UP000215335 UP000092444 UP000030765 UP000037069 UP000009192 UP000092443 UP000092445 UP000092460 UP000091820 UP000000803 UP000002282 UP000008711 UP000053825 UP000000311 UP000075809 UP000001070 UP000007801 UP000078541 UP000005205
Interpro
Gene 3D
ProteinModelPortal
H9IX89
A0A194QKJ2
A0A0N1I8H5
A0A2A4K9Q5
A0A2H1W1J9
A0A212FLR8
+ More
A0A2J7PJ95 A0A2J7PJ79 J9JY97 A0A1J1IIC7 A0A2H8TSM7 A0A182GP75 D6WR11 B0WY21 A0A1B0CXC2 Q16IL9 A0A2S2PV88 A0A1W4WYI0 A0A1B0EZK3 U4UEQ8 N6U904 K7J115 A0A1Y1JX99 A0A0K8TSD0 A0A1B6C1M9 A0A3Q0IIN9 A0A182IY11 A0A1I8MY57 B4NEP5 A0A0M4EYA8 A0A182RNQ2 T1PAQ8 A0A0C9REP4 A0A1I8Q5S1 A0A1A9VSZ3 A0A232F5K6 A0A1B0G9T9 A0A084VRD0 A0A1Y1JU48 A0A1B6KVE5 A0A0L0C524 B4L8U1 A0A2L2Y635 A0A1A9YM76 A0A1B6FTU9 A0A034WGL4 A0A1B0A1V0 D3TR07 A0A0K8U891 A0A1B0BII4 A0A1A9W1Z3 W8C0D0 Q8SXX0 B4PX89 B3NXH9 A0A0L7QQH4 E1ZYV8 A0A310SIF7 A0A151XCA9 B4JNV4 B3M5F7 A0A195F2B4 A0A158NFT7
A0A2J7PJ95 A0A2J7PJ79 J9JY97 A0A1J1IIC7 A0A2H8TSM7 A0A182GP75 D6WR11 B0WY21 A0A1B0CXC2 Q16IL9 A0A2S2PV88 A0A1W4WYI0 A0A1B0EZK3 U4UEQ8 N6U904 K7J115 A0A1Y1JX99 A0A0K8TSD0 A0A1B6C1M9 A0A3Q0IIN9 A0A182IY11 A0A1I8MY57 B4NEP5 A0A0M4EYA8 A0A182RNQ2 T1PAQ8 A0A0C9REP4 A0A1I8Q5S1 A0A1A9VSZ3 A0A232F5K6 A0A1B0G9T9 A0A084VRD0 A0A1Y1JU48 A0A1B6KVE5 A0A0L0C524 B4L8U1 A0A2L2Y635 A0A1A9YM76 A0A1B6FTU9 A0A034WGL4 A0A1B0A1V0 D3TR07 A0A0K8U891 A0A1B0BII4 A0A1A9W1Z3 W8C0D0 Q8SXX0 B4PX89 B3NXH9 A0A0L7QQH4 E1ZYV8 A0A310SIF7 A0A151XCA9 B4JNV4 B3M5F7 A0A195F2B4 A0A158NFT7
PDB
2E7G
E-value=2.47508e-11,
Score=165
Ontologies
GO
PANTHER
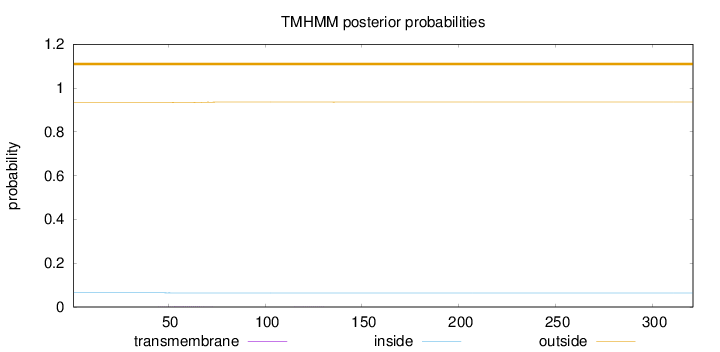
Topology
Length:
321
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03264
Exp number, first 60 AAs:
0.01718
Total prob of N-in:
0.06542
outside
1 - 321
Population Genetic Test Statistics
Pi
186.699525
Theta
24.908477
Tajima's D
0.360449
CLR
0.449304
CSRT
0.479826008699565
Interpretation
Uncertain
