Gene
KWMTBOMO11403
Pre Gene Modal
BGIBMGA001949
Annotation
PREDICTED:_retinoblastoma-like_protein_1_isoform_X2_[Papilio_xuthus]
Full name
Retinoblastoma-like protein 1
Alternative Name
107 kDa retinoblastoma-associated protein
pRb1
pRb1
Location in the cell
Nuclear Reliability : 3.886
Sequence
CDS
ATGCCCAAATCAGACGAAAACGAAGATTGGATTGAGAAGATGGAGGATCTTTGTTCTAATTTAAACGTCGATTCATCAGCTGCGAAGAAATCGAAAGATTCTTTTTCAGAAATAAAACGAAATTATATACTAGATGGTGATGAATTACATTGGATGGCATGCGCATTGTATGTGGCTTGTAGGACATCGGTCACTCCTACAGTGCAAACAGGAAAAGTGGTGGAAGGGAACTGTGTCAGCCTGACAAAACTTTTGAGATTATGTAATCTAAGCTTAATTCAGTTCTTCATCAAAATAAAAAACTGGATGGAGATGGCCTCAATGTCTACAGATTTTAAAGAAAGGATATCTCGTTTGGAACACAAGTTTGCAGTCTCTTCAGTACTCTTCAGGAAGTTCCAACCGATTTTCATGGAACTGTTTGTTGGGTTAACAAATGAGCCTGTAAAACAAGTCTCGAAGAGAAGACCAAAACTGCAACCATGTTCAACAAATGCCTTGTTTGAGTTTACATGGTGTTTGTACATTTGTGTGAAAGGAGAATTCCACAATTCTGCTAATGACTTAGTAGATATGTATCACATATTACTATCATGTTTGGATTATATATTTGCCAATGCATTTATGGCCCGAAGGGTAGACATTATTAATCCTGAATTTAAAGGATTACCTACAGATTGGACTAAAAATGATTTCAAAATGCCAAAAACTCCTCCATGTGTAATTTCTACTTTATGTGAGATCAAGGATGGCTTATCGAAAGAGGCAACCACTATGAAAGAATATAGCTGGAAACCTGTTATTGATTCTTTTTTTGAAAAAGGGATACTGAAAGGCAATAGTGAGCCTACGCTTGGAATTCTAGATTTGGGAGTGTTTGATGTAAATCTTAAATCACTGAACAACTTGTATGAAACATATGTTCTCAGTGTAGGAGAATTTGATGAAAGGATTTTTTTAGGCGAACAAGCAAATGAACAAATTGGAATGAAGAACAAAGTGTCCGGTGATGAGATCTCAGAAGTAATAGCTAATTTCGGTCCTAGCGGTCGTGCGTGCCCGGACACGCCGCTGACGGGGCGGCGCTACCTGTCGCGCCGCAGCGAGGAGCTGACGCCCGTGTCGGAGGCCAAGAACAGCCTGGCGCGCCTCGCCGCCACGCTGCGACACGCGCGCCCCCAACCCTCGCCCACGCTGCTCAGGCTCTTCACGGAATGCGGCGTAACTGATGAAATGATAAATGCAAAACTAATAAAACCCTGCAATGGTTGGATGGAACAATTTGGGGCCAGCTTGAAGGAAGCACACAATACCCCGCATAGTGAGACAATTTCTATACGCTGTAATATGGTCACATGTTTATATTATAAGGTGTTTGAACATATCATCAAAGAGGAACATAGGAAAAAGCCACAGGTTTCATTGCAGATGCTTCTTTCACAAGAAACATACCAGCTAACTGTATATGCGTGCTGTACGGAGATTGTCCTACATGCCTATGGAGTGAATTCATTCAAGTTTCCCAGAGTGCTGCAGATATTCAAGCTGAGCGCATTTCATTTCTATAAGATTATTGAACTAGTTGTACAGGCCGTGGTCGATAAGCTTAGTCGAGACGTCATCAAACATTTGAATGCAGTCGAAGAAGAAGTACTAGAGTCTCTGGTGTGGACTTCTGACAGTCCTCTGTGGGACCAACTCAGTAAAACGCCAGTGCCAGCCTCTGCCGACGTCTCCGTACAAGATTCACCTTTTCGAAGAACTAATGGTCTACAATCACCTGTCTCTACAATAGACCGCTTCATGTCACCTATGGCTGAACAAGCCAAGAAGCAATTATTCAAAGACCCTATTAAACCCGGCCAGTCACTTTTAGTACAAGCAAACACAACACCAGTGAAAGAGCCAGCAGTGTCTGCTAATACATCACAACCATCACAAAGTACTTCAAGTTGTGAGACCACGCCTACATCGACACCCAAAAAAAACAATTCATTAATATTGTTTTTTAGGAAGTTTTACAGTTTAGCGGTAGTACGTATGAACGATCTGTGCACGAGATTGCGACTGACTGACGACGAATTAAAACGCAAAATTTGGACTTGTCTTGAGTACTCAATAATGCATCAAACACATCTAATGAAAGACAGACATCTAGATCAGATACTTATGTGCTCTGTTTATGTTATATGTAAGGTTTCAAATAATCCGTCTAGTAATCAAGTGGAGAAAACATTTGCAGAAATCATGCGATGTTACAGACAACGCCCTTTAGCGGATAATCATGTTTATAGATCTGTTTTAATCAAACAAAGCACAGACGAGAGTTCACCAGAAAGAGGAGATTTGATTAACTTTTATAATAAAGTTTATGTGCAATGTATGCAGAATTTTGCACTTAGATTTTCTGGACGACACAGGGATGAGTGCAGCCTGTCTCCGCTGCCGACGGGGCGGTGTGAGGCGCGCTCCCCGGCCGGCACGCGCGTGTCCGAGCGACATCAGCTCTACGTCAAGCCGCTCACCGACCCGCCGCCGCAACACCACCACCTCACCTACAGATTCAGCCGCAGCCCCGCCAAGGACCTACAGGCGATCAACAGTTTGGTCTGGTGCGACACAGGACTGGGGTCGGGCGTAGGCCTGAAGCGTGCGCTGGAAGGAGCGTTGGACAGCGACCCGAATAAGCGAAACAGAGCCGCACCCGCCGTAGCCAGAAAGCTGCATCATCTACTATCTGACAGGCAGGCTGTCTAG
Protein
MPKSDENEDWIEKMEDLCSNLNVDSSAAKKSKDSFSEIKRNYILDGDELHWMACALYVACRTSVTPTVQTGKVVEGNCVSLTKLLRLCNLSLIQFFIKIKNWMEMASMSTDFKERISRLEHKFAVSSVLFRKFQPIFMELFVGLTNEPVKQVSKRRPKLQPCSTNALFEFTWCLYICVKGEFHNSANDLVDMYHILLSCLDYIFANAFMARRVDIINPEFKGLPTDWTKNDFKMPKTPPCVISTLCEIKDGLSKEATTMKEYSWKPVIDSFFEKGILKGNSEPTLGILDLGVFDVNLKSLNNLYETYVLSVGEFDERIFLGEQANEQIGMKNKVSGDEISEVIANFGPSGRACPDTPLTGRRYLSRRSEELTPVSEAKNSLARLAATLRHARPQPSPTLLRLFTECGVTDEMINAKLIKPCNGWMEQFGASLKEAHNTPHSETISIRCNMVTCLYYKVFEHIIKEEHRKKPQVSLQMLLSQETYQLTVYACCTEIVLHAYGVNSFKFPRVLQIFKLSAFHFYKIIELVVQAVVDKLSRDVIKHLNAVEEEVLESLVWTSDSPLWDQLSKTPVPASADVSVQDSPFRRTNGLQSPVSTIDRFMSPMAEQAKKQLFKDPIKPGQSLLVQANTTPVKEPAVSANTSQPSQSTSSCETTPTSTPKKNNSLILFFRKFYSLAVVRMNDLCTRLRLTDDELKRKIWTCLEYSIMHQTHLMKDRHLDQILMCSVYVICKVSNNPSSNQVEKTFAEIMRCYRQRPLADNHVYRSVLIKQSTDESSPERGDLINFYNKVYVQCMQNFALRFSGRHRDECSLSPLPTGRCEARSPAGTRVSERHQLYVKPLTDPPPQHHHLTYRFSRSPAKDLQAINSLVWCDTGLGSGVGLKRALEGALDSDPNKRNRAAPAVARKLHHLLSDRQAV
Summary
Description
Key regulator of entry into cell division. Directly involved in heterochromatin formation by maintaining overall chromatin structure and, in particular, that of constitutive heterochromatin by stabilizing histone methylation. Recruits and targets histone methyltransferases KMT5B and KMT5C, leading to epigenetic transcriptional repression. Controls histone H4 'Lys-20' trimethylation. Probably acts as a transcription repressor by recruiting chromatin-modifying enzymes to promoters. Potent inhibitor of E2F-mediated trans-activation. Forms a complex with adenovirus E1A and with SV40 large T antigen. May bind and modulate functionally certain cellular proteins with which T and E1A compete for pocket binding. May act as a tumor suppressor.
Subunit
Interacts with AATF. Interacts with KDM5A. Component of the DREAM complex (also named LINC complex) at least composed of E2F4, E2F5, LIN9, LIN37, LIN52, LIN54, MYBL1, MYBL2, RBL1, RBL2, RBBP4, TFDP1 and TFDP2. The complex exists in quiescent cells where it represses cell cycle-dependent genes. It dissociates in S phase when LIN9, LIN37, LIN52 and LIN54 form a subcomplex that binds to MYBL2 (By similarity). Interacts with KMT5B, KMT5C and USP4.
Similarity
Belongs to the retinoblastoma protein (RB) family.
Keywords
Alternative splicing
Cell cycle
Chromatin regulator
Complete proteome
Nucleus
Phosphoprotein
Reference proteome
Repressor
Transcription
Transcription regulation
Tumor suppressor
Feature
chain Retinoblastoma-like protein 1
splice variant In isoform Short.
splice variant In isoform Short.
Uniprot
A0A2A4KAP4
A0A194QES0
A0A0N0PBV8
A0A1B6KSV5
A0A0N0BD75
A0A067QPM2
+ More
A0A2A3EA33 A0A0C9S1V4 A0A088A345 A0A195F2H7 E2AA07 E9IWV1 A0A2J7PJ76 A0A1S3I9G5 A0A0X9C1Y3 V3ZXN9 K9J3F9 K7J2L5 A0A3P8RLF2 T1J484 A0A3P8Y5E5 A0A3Q0RN28 A0A3P8ZZU8 A0A3Q1C1I5 A0A3B1JF35 A0A2S2QXQ9 A0A3B4WX80 A0A1W4WTH2 A0A2H8TIK0 A0A336M5G2 A0A3Q2U3X7 A0A1Y1LA14 A0A1Y1LC63 A0A210R2R8 T1K8P0 A0A0K2TLX7 A0A1Y1LDR3 A0A1Y1LHI5 A0A2Y9KN45 A0A2R7WUQ8 A0A1Q3FAJ9 A0A287AAW9 G3TP05 A0A1L8DXE0 A0A0A1WQT9 A0A034WRH1 W8CCW6 H0XAP8 U4UFI4 N6U3F6 A0A2Y9QML1 A0A2Y9DA13 Q3UI58 A0A2J8MKW3 L5JXH6 F1SEM0 A0A2J8MKW4 A0A2Y9IP75 A0A286ZJW1 A0A340WDZ0 A0A2Y9IRQ4 A0A3M6UCC8 A0A287AEE7 A0A2K5URJ7 H9ZBY8 A0A2Y9PMP4 A0A2Y9T7Q3 A0A3Q2HLF0 A0A2K5DHS4 C3XU15 F7IJ08 A0A341BB53 A0A096NSC8 A0A3Q7PE55 A0A2Y9GZE8 F7CLV0 Q64701 A0A3Q7T7T3 Q6PAR4 A0A2U3XQM1 A0A2R9BWX0 F6X9R7 A0A2J8MKX3 A0A2U4CLS0 A0A384B0P2 A0A1U7RD35 G3QRW9 A0A2K6CV71 H2QKB0 A0A2R9BNQ5 A0A2C9K1J5
A0A2A3EA33 A0A0C9S1V4 A0A088A345 A0A195F2H7 E2AA07 E9IWV1 A0A2J7PJ76 A0A1S3I9G5 A0A0X9C1Y3 V3ZXN9 K9J3F9 K7J2L5 A0A3P8RLF2 T1J484 A0A3P8Y5E5 A0A3Q0RN28 A0A3P8ZZU8 A0A3Q1C1I5 A0A3B1JF35 A0A2S2QXQ9 A0A3B4WX80 A0A1W4WTH2 A0A2H8TIK0 A0A336M5G2 A0A3Q2U3X7 A0A1Y1LA14 A0A1Y1LC63 A0A210R2R8 T1K8P0 A0A0K2TLX7 A0A1Y1LDR3 A0A1Y1LHI5 A0A2Y9KN45 A0A2R7WUQ8 A0A1Q3FAJ9 A0A287AAW9 G3TP05 A0A1L8DXE0 A0A0A1WQT9 A0A034WRH1 W8CCW6 H0XAP8 U4UFI4 N6U3F6 A0A2Y9QML1 A0A2Y9DA13 Q3UI58 A0A2J8MKW3 L5JXH6 F1SEM0 A0A2J8MKW4 A0A2Y9IP75 A0A286ZJW1 A0A340WDZ0 A0A2Y9IRQ4 A0A3M6UCC8 A0A287AEE7 A0A2K5URJ7 H9ZBY8 A0A2Y9PMP4 A0A2Y9T7Q3 A0A3Q2HLF0 A0A2K5DHS4 C3XU15 F7IJ08 A0A341BB53 A0A096NSC8 A0A3Q7PE55 A0A2Y9GZE8 F7CLV0 Q64701 A0A3Q7T7T3 Q6PAR4 A0A2U3XQM1 A0A2R9BWX0 F6X9R7 A0A2J8MKX3 A0A2U4CLS0 A0A384B0P2 A0A1U7RD35 G3QRW9 A0A2K6CV71 H2QKB0 A0A2R9BNQ5 A0A2C9K1J5
Pubmed
26354079
24845553
20798317
21282665
23254933
20075255
+ More
25069045 25329095 15592404 28004739 28812685 25830018 25348373 24495485 23537049 10349636 11042159 11076861 11217851 12466851 16141073 23258410 30382153 25319552 19892987 18563158 17431167 8661722 7490090 16141072 19468303 11571651 15750587 17242355 21183079 15489334 22722832 22398555 16136131 15562597
25069045 25329095 15592404 28004739 28812685 25830018 25348373 24495485 23537049 10349636 11042159 11076861 11217851 12466851 16141073 23258410 30382153 25319552 19892987 18563158 17431167 8661722 7490090 16141072 19468303 11571651 15750587 17242355 21183079 15489334 22722832 22398555 16136131 15562597
EMBL
NWSH01000016
PCG80810.1
KQ459053
KPJ03977.1
KQ460772
KPJ12464.1
+ More
GEBQ01025448 JAT14529.1 KQ435878 KOX70054.1 KK853091 KDR11553.1 KZ288322 PBC28152.1 GBYB01014776 JAG84543.1 KQ981864 KYN34294.1 GL437990 EFN69736.1 GL766614 EFZ14956.1 NEVH01024950 PNF16395.1 KP640620 ALB00261.1 KB201489 ESO96313.1 GABZ01003951 JAA49574.1 JH431841 GGMS01013343 MBY82546.1 GFXV01002026 MBW13831.1 UFQT01000185 SSX21268.1 AADN05000479 GEZM01061429 JAV70492.1 GEZM01061433 JAV70488.1 NEDP02000731 OWF55242.1 CAEY01001879 HACA01009548 CDW26909.1 GEZM01061431 JAV70490.1 GEZM01061434 JAV70487.1 KK855619 PTY23299.1 GFDL01010496 JAV24549.1 AEMK02000106 GFDF01003022 JAV11062.1 GBXI01013271 JAD01021.1 GAKP01002172 JAC56780.1 GAMC01002981 JAC03575.1 AAQR03049853 AAQR03049854 AAQR03049855 AAQR03049856 AAQR03049857 AAQR03049858 AAQR03049859 AAQR03049860 AAQR03049861 AAQR03049862 KB632333 ERL92744.1 APGK01051096 KB741181 ENN73092.1 AK147064 BAE27648.1 NBAG03000253 PNI60164.1 KB031072 ELK04164.1 PNI60163.1 RCHS01001807 RMX51301.1 AQIA01008744 AQIA01008745 AQIA01008746 AQIA01008747 AQIA01008748 AQIA01008749 JU476650 AFH33454.1 GG666464 EEN68389.1 AHZZ02000391 AHZZ02000392 JSUE03004419 U33320 U27177 U27178 AK156055 AL669828 BC060124 AAH60124.1 AJFE02112947 AJFE02112948 AJFE02112949 AJFE02112950 AJFE02112951 AJFE02112952 AJFE02112953 AJFE02112954 AJFE02112955 AJFE02112956 PNI60162.1 CABD030116688 CABD030116689 CABD030116690 CABD030116691 CABD030116692 CABD030116693 CABD030116694 CABD030116695 CABD030116696 CABD030116697 AACZ04052783 GABC01010197 GABF01009275 GABF01009274 GABF01009273 GABD01007608 GABD01007607 GABE01009801 JAA01141.1 JAA12870.1 JAA25492.1 JAA34938.1 PNI60161.1
GEBQ01025448 JAT14529.1 KQ435878 KOX70054.1 KK853091 KDR11553.1 KZ288322 PBC28152.1 GBYB01014776 JAG84543.1 KQ981864 KYN34294.1 GL437990 EFN69736.1 GL766614 EFZ14956.1 NEVH01024950 PNF16395.1 KP640620 ALB00261.1 KB201489 ESO96313.1 GABZ01003951 JAA49574.1 JH431841 GGMS01013343 MBY82546.1 GFXV01002026 MBW13831.1 UFQT01000185 SSX21268.1 AADN05000479 GEZM01061429 JAV70492.1 GEZM01061433 JAV70488.1 NEDP02000731 OWF55242.1 CAEY01001879 HACA01009548 CDW26909.1 GEZM01061431 JAV70490.1 GEZM01061434 JAV70487.1 KK855619 PTY23299.1 GFDL01010496 JAV24549.1 AEMK02000106 GFDF01003022 JAV11062.1 GBXI01013271 JAD01021.1 GAKP01002172 JAC56780.1 GAMC01002981 JAC03575.1 AAQR03049853 AAQR03049854 AAQR03049855 AAQR03049856 AAQR03049857 AAQR03049858 AAQR03049859 AAQR03049860 AAQR03049861 AAQR03049862 KB632333 ERL92744.1 APGK01051096 KB741181 ENN73092.1 AK147064 BAE27648.1 NBAG03000253 PNI60164.1 KB031072 ELK04164.1 PNI60163.1 RCHS01001807 RMX51301.1 AQIA01008744 AQIA01008745 AQIA01008746 AQIA01008747 AQIA01008748 AQIA01008749 JU476650 AFH33454.1 GG666464 EEN68389.1 AHZZ02000391 AHZZ02000392 JSUE03004419 U33320 U27177 U27178 AK156055 AL669828 BC060124 AAH60124.1 AJFE02112947 AJFE02112948 AJFE02112949 AJFE02112950 AJFE02112951 AJFE02112952 AJFE02112953 AJFE02112954 AJFE02112955 AJFE02112956 PNI60162.1 CABD030116688 CABD030116689 CABD030116690 CABD030116691 CABD030116692 CABD030116693 CABD030116694 CABD030116695 CABD030116696 CABD030116697 AACZ04052783 GABC01010197 GABF01009275 GABF01009274 GABF01009273 GABD01007608 GABD01007607 GABE01009801 JAA01141.1 JAA12870.1 JAA25492.1 JAA34938.1 PNI60161.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000053105
UP000027135
UP000242457
+ More
UP000005203 UP000078541 UP000000311 UP000235965 UP000085678 UP000030746 UP000002358 UP000265080 UP000265140 UP000261340 UP000257160 UP000018467 UP000261360 UP000192223 UP000000539 UP000242188 UP000015104 UP000248482 UP000008227 UP000007646 UP000005225 UP000030742 UP000019118 UP000248480 UP000010552 UP000265300 UP000275408 UP000233100 UP000248483 UP000248484 UP000002281 UP000233020 UP000001554 UP000008225 UP000252040 UP000028761 UP000286641 UP000248481 UP000006718 UP000000589 UP000286640 UP000245341 UP000240080 UP000245320 UP000261681 UP000189706 UP000001519 UP000233120 UP000002277 UP000076420
UP000005203 UP000078541 UP000000311 UP000235965 UP000085678 UP000030746 UP000002358 UP000265080 UP000265140 UP000261340 UP000257160 UP000018467 UP000261360 UP000192223 UP000000539 UP000242188 UP000015104 UP000248482 UP000008227 UP000007646 UP000005225 UP000030742 UP000019118 UP000248480 UP000010552 UP000265300 UP000275408 UP000233100 UP000248483 UP000248484 UP000002281 UP000233020 UP000001554 UP000008225 UP000252040 UP000028761 UP000286641 UP000248481 UP000006718 UP000000589 UP000286640 UP000245341 UP000240080 UP000245320 UP000261681 UP000189706 UP000001519 UP000233120 UP000002277 UP000076420
Interpro
SUPFAM
SSF47954
SSF47954
CDD
ProteinModelPortal
A0A2A4KAP4
A0A194QES0
A0A0N0PBV8
A0A1B6KSV5
A0A0N0BD75
A0A067QPM2
+ More
A0A2A3EA33 A0A0C9S1V4 A0A088A345 A0A195F2H7 E2AA07 E9IWV1 A0A2J7PJ76 A0A1S3I9G5 A0A0X9C1Y3 V3ZXN9 K9J3F9 K7J2L5 A0A3P8RLF2 T1J484 A0A3P8Y5E5 A0A3Q0RN28 A0A3P8ZZU8 A0A3Q1C1I5 A0A3B1JF35 A0A2S2QXQ9 A0A3B4WX80 A0A1W4WTH2 A0A2H8TIK0 A0A336M5G2 A0A3Q2U3X7 A0A1Y1LA14 A0A1Y1LC63 A0A210R2R8 T1K8P0 A0A0K2TLX7 A0A1Y1LDR3 A0A1Y1LHI5 A0A2Y9KN45 A0A2R7WUQ8 A0A1Q3FAJ9 A0A287AAW9 G3TP05 A0A1L8DXE0 A0A0A1WQT9 A0A034WRH1 W8CCW6 H0XAP8 U4UFI4 N6U3F6 A0A2Y9QML1 A0A2Y9DA13 Q3UI58 A0A2J8MKW3 L5JXH6 F1SEM0 A0A2J8MKW4 A0A2Y9IP75 A0A286ZJW1 A0A340WDZ0 A0A2Y9IRQ4 A0A3M6UCC8 A0A287AEE7 A0A2K5URJ7 H9ZBY8 A0A2Y9PMP4 A0A2Y9T7Q3 A0A3Q2HLF0 A0A2K5DHS4 C3XU15 F7IJ08 A0A341BB53 A0A096NSC8 A0A3Q7PE55 A0A2Y9GZE8 F7CLV0 Q64701 A0A3Q7T7T3 Q6PAR4 A0A2U3XQM1 A0A2R9BWX0 F6X9R7 A0A2J8MKX3 A0A2U4CLS0 A0A384B0P2 A0A1U7RD35 G3QRW9 A0A2K6CV71 H2QKB0 A0A2R9BNQ5 A0A2C9K1J5
A0A2A3EA33 A0A0C9S1V4 A0A088A345 A0A195F2H7 E2AA07 E9IWV1 A0A2J7PJ76 A0A1S3I9G5 A0A0X9C1Y3 V3ZXN9 K9J3F9 K7J2L5 A0A3P8RLF2 T1J484 A0A3P8Y5E5 A0A3Q0RN28 A0A3P8ZZU8 A0A3Q1C1I5 A0A3B1JF35 A0A2S2QXQ9 A0A3B4WX80 A0A1W4WTH2 A0A2H8TIK0 A0A336M5G2 A0A3Q2U3X7 A0A1Y1LA14 A0A1Y1LC63 A0A210R2R8 T1K8P0 A0A0K2TLX7 A0A1Y1LDR3 A0A1Y1LHI5 A0A2Y9KN45 A0A2R7WUQ8 A0A1Q3FAJ9 A0A287AAW9 G3TP05 A0A1L8DXE0 A0A0A1WQT9 A0A034WRH1 W8CCW6 H0XAP8 U4UFI4 N6U3F6 A0A2Y9QML1 A0A2Y9DA13 Q3UI58 A0A2J8MKW3 L5JXH6 F1SEM0 A0A2J8MKW4 A0A2Y9IP75 A0A286ZJW1 A0A340WDZ0 A0A2Y9IRQ4 A0A3M6UCC8 A0A287AEE7 A0A2K5URJ7 H9ZBY8 A0A2Y9PMP4 A0A2Y9T7Q3 A0A3Q2HLF0 A0A2K5DHS4 C3XU15 F7IJ08 A0A341BB53 A0A096NSC8 A0A3Q7PE55 A0A2Y9GZE8 F7CLV0 Q64701 A0A3Q7T7T3 Q6PAR4 A0A2U3XQM1 A0A2R9BWX0 F6X9R7 A0A2J8MKX3 A0A2U4CLS0 A0A384B0P2 A0A1U7RD35 G3QRW9 A0A2K6CV71 H2QKB0 A0A2R9BNQ5 A0A2C9K1J5
PDB
4ELL
E-value=8.93549e-43,
Score=440
Ontologies
GO
GO:0051726
GO:0005634
GO:0006357
GO:0005667
GO:0051302
GO:0016021
GO:0005654
GO:0043550
GO:1990841
GO:0000122
GO:2000773
GO:0008134
GO:0010629
GO:2000134
GO:0030154
GO:0001012
GO:0001102
GO:0007346
GO:0000785
GO:0006325
GO:0007049
GO:0006313
GO:0007018
GO:0030286
GO:0046034
GO:0032559
GO:0004812
GO:0006418
GO:0003824
PANTHER
Topology
Subcellular location
Nucleus
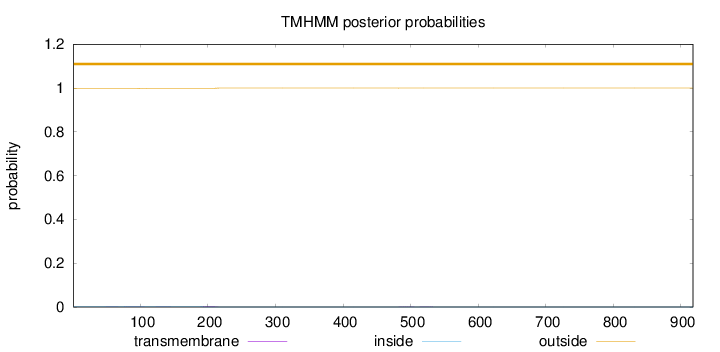
Length:
918
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0751199999999997
Exp number, first 60 AAs:
0.00221
Total prob of N-in:
0.00311
outside
1 - 918
Population Genetic Test Statistics
Pi
177.435306
Theta
164.278261
Tajima's D
-0.453147
CLR
1.168116
CSRT
0.254287285635718
Interpretation
Uncertain
