Gene
KWMTBOMO11402
Pre Gene Modal
BGIBMGA001871
Annotation
PREDICTED:_SAGA-associated_factor_29_homolog_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.029 Mitochondrial Reliability : 1.624 Nuclear Reliability : 1.174
Sequence
CDS
ATGCCTCTCACAGCTGACGTTGCTGCACAACAAGTACAGGAAAGATTGAGGTCATTACACAGATTGATTTATGATATAGAAGCGGAGAGGTCTCGCAATGAACAGTGCATAGATGCCATACTTAGAGCTGAAAAATCTACAGGATCTCCTTCGAGTGCTGATGATTCATCAGGATCTTCCCAACAAGTTTCGCAGCTTAAAAGTTTATATAAATCGGGCTTAACAGCTGCCGAACAAGAGGAAAGACTACTTAGAGCTGCACTGTCAAGGATTTACGAAATACGTGCAATTAAAAATGAACGTCGAATTCAGGCTCGTCACGCGGGTAACAAGGAGACGATCGGTTGCGGGGCGCTGATGAAGCTGCTCCTGAGTGCCGCGCAAACGTTGCCGCTGCACGTGGGGCGCGCGGGGGAGCGGGCGCCGCCGCTGTGCGGAGCGGTGCCGGCCGACGCGGGGCACGTCGCTCGACCCGGGGACGCCGTCGCCGCGCTCGTGCGGGTCTCCGATAACGAGGACAACTGGATATTGGCCGAGGTGGTGAGCTGGTTGCCCGCTCAGGGGAAATACGAAGTGGACGACATCGACGAGGAGCAGAAGAATCGTCACGTGCTGAGCAAGCGCCGCGTCGTGCCGCTGCCGCTGATGCGAGCCGACCCTCGGACCGACGAACACGCTCTCTTCCCTAAAGGGCAGTAA
Protein
MPLTADVAAQQVQERLRSLHRLIYDIEAERSRNEQCIDAILRAEKSTGSPSSADDSSGSSQQVSQLKSLYKSGLTAAEQEERLLRAALSRIYEIRAIKNERRIQARHAGNKETIGCGALMKLLLSAAQTLPLHVGRAGERAPPLCGAVPADAGHVARPGDAVAALVRVSDNEDNWILAEVVSWLPAQGKYEVDDIDEEQKNRHVLSKRRVVPLPLMRADPRTDEHALFPKGQ
Summary
Uniprot
A0A194QEL4
A0A3S2P1B1
A0A0N1IF64
A0A2H1WRL7
A0A212EUH5
A0A2A4K9B2
+ More
H9IX88 A0A1B0DFB8 A0A1B0CVB9 A0A067QXF8 A0A0K8TL66 A0A1L8DX25 A0A2A3ESC7 A0A088A5K9 A0A232FA38 A0A182LBB5 Q7PQ24 A0A182HP75 Q16IF7 A0A182TKQ2 A0A154P8S1 A0A2J7PT46 A0A0L7R9J7 K7IR13 A0A182VBF6 A0A182GM64 E0VAG4 A0A195EJH7 A0A023EML2 A0A182JTN0 A0A182FCG1 A0A182WAA1 A0A182RTN5 U5ET93 E1ZY36 A0A2M4CN35 A0A195BEN3 A0A158NQT1 A0A182QPX8 T1PHJ8 B4KSJ5 A0A195FEC6 A0A026W745 A0A1I8N348 A0A182N816 A0A151WSF2 W5JVB0 A0A1Q3F8I3 A0A195CBF7 B4J7I1 D3TKV7 A0A1B6CIQ2 A0A1B0ACF3 B0WCB0 A0A182XJ87 A0A1A9UY52 A0A182JJ11 Q9W2I4 A0A336KFT2 A0A0L0C730 E9FTA9 B4LMG2 F4WI07 A0A0P5WZC7 A0A182MLN5 A0A0T6ATP2 B4I7N4 B4QG11 A0A1A9Y0F8 A0A1B0B662 A0A0P5XHA4 A0A0P4VS55 A0A0P5BD41 A0A1W4UAU8 A0A1I8Q9S7 A0A0K8V9L5 A0A034V8U9 A0A182YFL0 A0A023F0S9 A0A224XUZ8 T1GGY6 E9IPT0 A0A182PHC9 A0A0P6AI86 B4P9E3 B3NJL7 A0A0P5BBS0 A0A0V0G898 A0A0P5B5V3 A0A3B0J2U6 B3MBW6 Q291V3 A0A0M8ZYQ9 A0A1B0FQG4 A0A2P8YAR6 A0A0M4ECW6 A0A0A1X8Q0 A0A084W9L2 W8BBI2 A0A1A9WS62
H9IX88 A0A1B0DFB8 A0A1B0CVB9 A0A067QXF8 A0A0K8TL66 A0A1L8DX25 A0A2A3ESC7 A0A088A5K9 A0A232FA38 A0A182LBB5 Q7PQ24 A0A182HP75 Q16IF7 A0A182TKQ2 A0A154P8S1 A0A2J7PT46 A0A0L7R9J7 K7IR13 A0A182VBF6 A0A182GM64 E0VAG4 A0A195EJH7 A0A023EML2 A0A182JTN0 A0A182FCG1 A0A182WAA1 A0A182RTN5 U5ET93 E1ZY36 A0A2M4CN35 A0A195BEN3 A0A158NQT1 A0A182QPX8 T1PHJ8 B4KSJ5 A0A195FEC6 A0A026W745 A0A1I8N348 A0A182N816 A0A151WSF2 W5JVB0 A0A1Q3F8I3 A0A195CBF7 B4J7I1 D3TKV7 A0A1B6CIQ2 A0A1B0ACF3 B0WCB0 A0A182XJ87 A0A1A9UY52 A0A182JJ11 Q9W2I4 A0A336KFT2 A0A0L0C730 E9FTA9 B4LMG2 F4WI07 A0A0P5WZC7 A0A182MLN5 A0A0T6ATP2 B4I7N4 B4QG11 A0A1A9Y0F8 A0A1B0B662 A0A0P5XHA4 A0A0P4VS55 A0A0P5BD41 A0A1W4UAU8 A0A1I8Q9S7 A0A0K8V9L5 A0A034V8U9 A0A182YFL0 A0A023F0S9 A0A224XUZ8 T1GGY6 E9IPT0 A0A182PHC9 A0A0P6AI86 B4P9E3 B3NJL7 A0A0P5BBS0 A0A0V0G898 A0A0P5B5V3 A0A3B0J2U6 B3MBW6 Q291V3 A0A0M8ZYQ9 A0A1B0FQG4 A0A2P8YAR6 A0A0M4ECW6 A0A0A1X8Q0 A0A084W9L2 W8BBI2 A0A1A9WS62
Pubmed
26354079
22118469
19121390
24845553
26369729
28648823
+ More
20966253 12364791 14747013 17210077 17510324 20075255 26483478 20566863 24945155 20798317 21347285 17994087 24508170 30249741 25315136 20920257 23761445 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 21292972 21719571 22936249 27129103 25348373 25244985 25474469 21282665 17550304 15632085 29403074 25830018 24438588 24495485
20966253 12364791 14747013 17210077 17510324 20075255 26483478 20566863 24945155 20798317 21347285 17994087 24508170 30249741 25315136 20920257 23761445 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 21292972 21719571 22936249 27129103 25348373 25244985 25474469 21282665 17550304 15632085 29403074 25830018 24438588 24495485
EMBL
KQ459053
KPJ03978.1
RSAL01000004
RVE54537.1
KQ460772
KPJ12463.1
+ More
ODYU01010530 SOQ55703.1 AGBW02012379 OWR45150.1 NWSH01000016 PCG80811.1 BABH01018123 BABH01018124 AJVK01035194 AJVK01058655 AJWK01030406 KK853244 KDR09432.1 GDAI01002702 JAI14901.1 GFDF01003077 JAV11007.1 KZ288189 PBC34607.1 NNAY01000622 OXU27340.1 AAAB01008900 EAA09403.5 APCN01002922 CH478080 CH477632 CH477581 EAT34052.1 EAT38054.1 EAT38684.1 KQ434827 KZC07530.1 NEVH01021921 PNF19495.1 KQ414626 KOC67504.1 JXUM01073894 KQ562799 KXJ75106.1 DS235006 EEB10370.1 KQ978822 KYN28067.1 GAPW01003020 JAC10578.1 GANO01002030 JAB57841.1 GL435171 EFN73885.1 GGFL01002515 MBW66693.1 KQ976500 KYM83051.1 ADTU01023495 AXCN02000912 KA648297 AFP62926.1 CH933808 EDW09500.1 KQ981636 KYN38731.1 KK107372 QOIP01000004 EZA51808.1 RLU23197.1 KQ982773 KYQ50800.1 ADMH02000391 ETN66734.1 GFDL01011222 JAV23823.1 KQ978068 KYM97448.1 CH916367 EDW01105.1 EZ422044 ADD18467.1 GEDC01024075 GEDC01001155 JAS13223.1 JAS36143.1 DS231886 EDS43355.1 AE013599 BT030796 AAF46707.2 ABV82178.1 AHN56468.1 UFQS01000264 UFQT01000264 SSX02195.1 SSX22572.1 JRES01000827 KNC28046.1 GL732524 EFX89337.1 CH940648 EDW62057.1 GL888170 EGI66158.1 GDIQ01245554 GDIQ01186918 GDIP01079410 LRGB01003123 JAK06171.1 JAM24305.1 KZS04259.1 AXCM01000363 LJIG01022878 KRT78267.1 CH480824 EDW56609.1 CM000362 CM002911 EDX08047.1 KMY95499.1 JXJN01008987 GDIP01072115 JAM31600.1 GDKW01001183 JAI55412.1 GDIP01186202 JAJ37200.1 GDHF01016660 JAI35654.1 GAKP01020777 GAKP01020772 JAC38175.1 GBBI01004123 JAC14589.1 GFTR01004617 JAW11809.1 CAQQ02392450 GL764626 EFZ17401.1 GDIP01029011 JAM74704.1 CM000158 EDW91263.1 CH954179 EDV55440.1 GDIP01191995 JAJ31407.1 GECL01001769 JAP04355.1 GDIP01189650 JAJ33752.1 OUUW01000001 SPP73473.1 CH902619 EDV36137.1 CM000071 EAL25009.1 KQ435794 KOX73830.1 CCAG010009452 PYGN01000749 PSN41350.1 CP012524 ALC41665.1 GBXI01006795 JAD07497.1 ATLV01021831 KE525324 KFB46906.1 GAMC01019341 JAB87214.1
ODYU01010530 SOQ55703.1 AGBW02012379 OWR45150.1 NWSH01000016 PCG80811.1 BABH01018123 BABH01018124 AJVK01035194 AJVK01058655 AJWK01030406 KK853244 KDR09432.1 GDAI01002702 JAI14901.1 GFDF01003077 JAV11007.1 KZ288189 PBC34607.1 NNAY01000622 OXU27340.1 AAAB01008900 EAA09403.5 APCN01002922 CH478080 CH477632 CH477581 EAT34052.1 EAT38054.1 EAT38684.1 KQ434827 KZC07530.1 NEVH01021921 PNF19495.1 KQ414626 KOC67504.1 JXUM01073894 KQ562799 KXJ75106.1 DS235006 EEB10370.1 KQ978822 KYN28067.1 GAPW01003020 JAC10578.1 GANO01002030 JAB57841.1 GL435171 EFN73885.1 GGFL01002515 MBW66693.1 KQ976500 KYM83051.1 ADTU01023495 AXCN02000912 KA648297 AFP62926.1 CH933808 EDW09500.1 KQ981636 KYN38731.1 KK107372 QOIP01000004 EZA51808.1 RLU23197.1 KQ982773 KYQ50800.1 ADMH02000391 ETN66734.1 GFDL01011222 JAV23823.1 KQ978068 KYM97448.1 CH916367 EDW01105.1 EZ422044 ADD18467.1 GEDC01024075 GEDC01001155 JAS13223.1 JAS36143.1 DS231886 EDS43355.1 AE013599 BT030796 AAF46707.2 ABV82178.1 AHN56468.1 UFQS01000264 UFQT01000264 SSX02195.1 SSX22572.1 JRES01000827 KNC28046.1 GL732524 EFX89337.1 CH940648 EDW62057.1 GL888170 EGI66158.1 GDIQ01245554 GDIQ01186918 GDIP01079410 LRGB01003123 JAK06171.1 JAM24305.1 KZS04259.1 AXCM01000363 LJIG01022878 KRT78267.1 CH480824 EDW56609.1 CM000362 CM002911 EDX08047.1 KMY95499.1 JXJN01008987 GDIP01072115 JAM31600.1 GDKW01001183 JAI55412.1 GDIP01186202 JAJ37200.1 GDHF01016660 JAI35654.1 GAKP01020777 GAKP01020772 JAC38175.1 GBBI01004123 JAC14589.1 GFTR01004617 JAW11809.1 CAQQ02392450 GL764626 EFZ17401.1 GDIP01029011 JAM74704.1 CM000158 EDW91263.1 CH954179 EDV55440.1 GDIP01191995 JAJ31407.1 GECL01001769 JAP04355.1 GDIP01189650 JAJ33752.1 OUUW01000001 SPP73473.1 CH902619 EDV36137.1 CM000071 EAL25009.1 KQ435794 KOX73830.1 CCAG010009452 PYGN01000749 PSN41350.1 CP012524 ALC41665.1 GBXI01006795 JAD07497.1 ATLV01021831 KE525324 KFB46906.1 GAMC01019341 JAB87214.1
Proteomes
UP000053268
UP000283053
UP000053240
UP000007151
UP000218220
UP000005204
+ More
UP000092462 UP000092461 UP000027135 UP000242457 UP000005203 UP000215335 UP000075882 UP000007062 UP000075840 UP000008820 UP000075902 UP000076502 UP000235965 UP000053825 UP000002358 UP000075903 UP000069940 UP000249989 UP000009046 UP000078492 UP000075881 UP000069272 UP000075920 UP000075900 UP000000311 UP000078540 UP000005205 UP000075886 UP000009192 UP000078541 UP000053097 UP000279307 UP000095301 UP000075884 UP000075809 UP000000673 UP000078542 UP000001070 UP000092445 UP000002320 UP000076407 UP000078200 UP000075880 UP000000803 UP000037069 UP000000305 UP000008792 UP000007755 UP000076858 UP000075883 UP000001292 UP000000304 UP000092443 UP000092460 UP000192221 UP000095300 UP000076408 UP000015102 UP000075885 UP000002282 UP000008711 UP000268350 UP000007801 UP000001819 UP000053105 UP000092444 UP000245037 UP000092553 UP000030765 UP000091820
UP000092462 UP000092461 UP000027135 UP000242457 UP000005203 UP000215335 UP000075882 UP000007062 UP000075840 UP000008820 UP000075902 UP000076502 UP000235965 UP000053825 UP000002358 UP000075903 UP000069940 UP000249989 UP000009046 UP000078492 UP000075881 UP000069272 UP000075920 UP000075900 UP000000311 UP000078540 UP000005205 UP000075886 UP000009192 UP000078541 UP000053097 UP000279307 UP000095301 UP000075884 UP000075809 UP000000673 UP000078542 UP000001070 UP000092445 UP000002320 UP000076407 UP000078200 UP000075880 UP000000803 UP000037069 UP000000305 UP000008792 UP000007755 UP000076858 UP000075883 UP000001292 UP000000304 UP000092443 UP000092460 UP000192221 UP000095300 UP000076408 UP000015102 UP000075885 UP000002282 UP000008711 UP000268350 UP000007801 UP000001819 UP000053105 UP000092444 UP000245037 UP000092553 UP000030765 UP000091820
Pfam
PF07039 DUF1325
ProteinModelPortal
A0A194QEL4
A0A3S2P1B1
A0A0N1IF64
A0A2H1WRL7
A0A212EUH5
A0A2A4K9B2
+ More
H9IX88 A0A1B0DFB8 A0A1B0CVB9 A0A067QXF8 A0A0K8TL66 A0A1L8DX25 A0A2A3ESC7 A0A088A5K9 A0A232FA38 A0A182LBB5 Q7PQ24 A0A182HP75 Q16IF7 A0A182TKQ2 A0A154P8S1 A0A2J7PT46 A0A0L7R9J7 K7IR13 A0A182VBF6 A0A182GM64 E0VAG4 A0A195EJH7 A0A023EML2 A0A182JTN0 A0A182FCG1 A0A182WAA1 A0A182RTN5 U5ET93 E1ZY36 A0A2M4CN35 A0A195BEN3 A0A158NQT1 A0A182QPX8 T1PHJ8 B4KSJ5 A0A195FEC6 A0A026W745 A0A1I8N348 A0A182N816 A0A151WSF2 W5JVB0 A0A1Q3F8I3 A0A195CBF7 B4J7I1 D3TKV7 A0A1B6CIQ2 A0A1B0ACF3 B0WCB0 A0A182XJ87 A0A1A9UY52 A0A182JJ11 Q9W2I4 A0A336KFT2 A0A0L0C730 E9FTA9 B4LMG2 F4WI07 A0A0P5WZC7 A0A182MLN5 A0A0T6ATP2 B4I7N4 B4QG11 A0A1A9Y0F8 A0A1B0B662 A0A0P5XHA4 A0A0P4VS55 A0A0P5BD41 A0A1W4UAU8 A0A1I8Q9S7 A0A0K8V9L5 A0A034V8U9 A0A182YFL0 A0A023F0S9 A0A224XUZ8 T1GGY6 E9IPT0 A0A182PHC9 A0A0P6AI86 B4P9E3 B3NJL7 A0A0P5BBS0 A0A0V0G898 A0A0P5B5V3 A0A3B0J2U6 B3MBW6 Q291V3 A0A0M8ZYQ9 A0A1B0FQG4 A0A2P8YAR6 A0A0M4ECW6 A0A0A1X8Q0 A0A084W9L2 W8BBI2 A0A1A9WS62
H9IX88 A0A1B0DFB8 A0A1B0CVB9 A0A067QXF8 A0A0K8TL66 A0A1L8DX25 A0A2A3ESC7 A0A088A5K9 A0A232FA38 A0A182LBB5 Q7PQ24 A0A182HP75 Q16IF7 A0A182TKQ2 A0A154P8S1 A0A2J7PT46 A0A0L7R9J7 K7IR13 A0A182VBF6 A0A182GM64 E0VAG4 A0A195EJH7 A0A023EML2 A0A182JTN0 A0A182FCG1 A0A182WAA1 A0A182RTN5 U5ET93 E1ZY36 A0A2M4CN35 A0A195BEN3 A0A158NQT1 A0A182QPX8 T1PHJ8 B4KSJ5 A0A195FEC6 A0A026W745 A0A1I8N348 A0A182N816 A0A151WSF2 W5JVB0 A0A1Q3F8I3 A0A195CBF7 B4J7I1 D3TKV7 A0A1B6CIQ2 A0A1B0ACF3 B0WCB0 A0A182XJ87 A0A1A9UY52 A0A182JJ11 Q9W2I4 A0A336KFT2 A0A0L0C730 E9FTA9 B4LMG2 F4WI07 A0A0P5WZC7 A0A182MLN5 A0A0T6ATP2 B4I7N4 B4QG11 A0A1A9Y0F8 A0A1B0B662 A0A0P5XHA4 A0A0P4VS55 A0A0P5BD41 A0A1W4UAU8 A0A1I8Q9S7 A0A0K8V9L5 A0A034V8U9 A0A182YFL0 A0A023F0S9 A0A224XUZ8 T1GGY6 E9IPT0 A0A182PHC9 A0A0P6AI86 B4P9E3 B3NJL7 A0A0P5BBS0 A0A0V0G898 A0A0P5B5V3 A0A3B0J2U6 B3MBW6 Q291V3 A0A0M8ZYQ9 A0A1B0FQG4 A0A2P8YAR6 A0A0M4ECW6 A0A0A1X8Q0 A0A084W9L2 W8BBI2 A0A1A9WS62
PDB
5C0M
E-value=1.46008e-33,
Score=354
Ontologies
GO
PANTHER
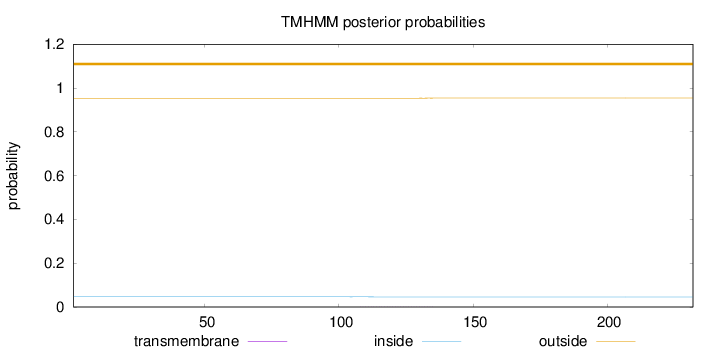
Topology
Length:
232
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02134
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04648
outside
1 - 232
Population Genetic Test Statistics
Pi
198.029457
Theta
151.092482
Tajima's D
0.879492
CLR
0.047146
CSRT
0.626418679066047
Interpretation
Uncertain
