Gene
KWMTBOMO11401 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001950
Annotation
PREDICTED:_polyadenylate_binding_protein_2_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.929
Sequence
CDS
ATGGCAGAAAATGATGATTTTTTAGACTCGATTGACAACAATCATTTGGAAGGAACTAACGGCTCAATGAATATGACTGACAATAGTCTCGGAGGCGGAGATGAAGCGGCCGGCGGAGAAGCTCCTGACCTTGCAGCTATCAAGGCCCGAGTGCGGGAGATGGAAGAAGAAGCTGAGAAACTGAAACAAATGCAGACTGAAGTTGACAAGCAAATGAGTATGGGAAGCCCACCAGGACTAACCAGTCCATTAAACATGTCTATCGAAGAAAAAATAGAAGCAGACAATAGATCAGTATATGTTGGTAATGTTGATTATGGAGCCACGGCTGAGGAGCTGGAGCAGCACTTTCATGGTTGTGGATCTATTAACAGGGTTACAATATTATGCAATAAGTATGATGGCCACCCAAAAGGTTTTGCCTACATCGAATTTGGCGACAAGGATAGTGTCCAGACTGCAATGGCTATGGATGAATCATTATTTAGAGGAAGGCAAATAAAGGTGATGCCGAAGCGTACGAACAAGCCGGGTCTGTCGTCTACGAACCGCCCGCCGCGCACGATGCGCGGGCGCGGGCGCGGCTACCGCGGCGGCCTCGCGCCTTACTACACGGGCTTCCGGCCCCCGCGACGCGGCAGAGTCTTCAAGCGCGGCTCGTACTACTCCCCGTACTGA
Protein
MAENDDFLDSIDNNHLEGTNGSMNMTDNSLGGGDEAAGGEAPDLAAIKARVREMEEEAEKLKQMQTEVDKQMSMGSPPGLTSPLNMSIEEKIEADNRSVYVGNVDYGATAEELEQHFHGCGSINRVTILCNKYDGHPKGFAYIEFGDKDSVQTAMAMDESLFRGRQIKVMPKRTNKPGLSSTNRPPRTMRGRGRGYRGGLAPYYTGFRPPRRGRVFKRGSYYSPY
Summary
Uniprot
Q0ZAL2
A0A0N0PC56
A0A3S2LIF7
A0A2A4KAX8
I4DRD6
I4DKS0
+ More
A0A212EUL3 A0A194QEY6 S4PB04 H9IXG7 A0A2H1VGH8 A0A1W4WYD2 A0A1Y1LEM5 D6WR37 J3JW69 A0A1Y1LEJ1 A0A1B6EB13 A0A0L7R948 A0A026W8N4 E2AU35 A0A2A3EJC1 V9IJU3 A0A087ZZC6 F4WGU3 A0A151JUW6 A0A158NFG4 A0A0J7NW30 A0A1B6LN98 A0A1B6HV38 A0A1B6FNI8 A0A232F094 A0A0P4WCG7 E0VAG5 A0A3Q0IR86 A0A0V0G5F6 A0A023F9K9 A0A069DPF5 A0A0P4VTH3 R4FLS7 A0A224XY24 R4WRH0 A0A170VQN7 A0A0B7AXY8 A0A1L8DH27 G1P0Z9 A0A1L8DH72 A0A0P5WSI9 A0A3S1BJ78 A0A0P5FLT9 A0A0K2ULU7 A0A0P5BTJ8 A0A0P4YRM3 A0A0P4Y821 A0A0P5K4N8 A0A0P5BMM0 A0A0K2UKQ9 A0A1J1IT08
A0A212EUL3 A0A194QEY6 S4PB04 H9IXG7 A0A2H1VGH8 A0A1W4WYD2 A0A1Y1LEM5 D6WR37 J3JW69 A0A1Y1LEJ1 A0A1B6EB13 A0A0L7R948 A0A026W8N4 E2AU35 A0A2A3EJC1 V9IJU3 A0A087ZZC6 F4WGU3 A0A151JUW6 A0A158NFG4 A0A0J7NW30 A0A1B6LN98 A0A1B6HV38 A0A1B6FNI8 A0A232F094 A0A0P4WCG7 E0VAG5 A0A3Q0IR86 A0A0V0G5F6 A0A023F9K9 A0A069DPF5 A0A0P4VTH3 R4FLS7 A0A224XY24 R4WRH0 A0A170VQN7 A0A0B7AXY8 A0A1L8DH27 G1P0Z9 A0A1L8DH72 A0A0P5WSI9 A0A3S1BJ78 A0A0P5FLT9 A0A0K2ULU7 A0A0P5BTJ8 A0A0P4YRM3 A0A0P4Y821 A0A0P5K4N8 A0A0P5BMM0 A0A0K2UKQ9 A0A1J1IT08
Pubmed
EMBL
DQ648527
ABG43002.1
KQ460772
KPJ12462.1
RSAL01000004
RVE54538.1
+ More
NWSH01000016 PCG80812.1 AK405074 BAM20476.1 AK401888 BAM18510.1 AGBW02012379 OWR45151.1 KQ459053 KPJ03979.1 GAIX01006192 JAA86368.1 BABH01018125 ODYU01002448 SOQ39945.1 GEZM01057951 JAV72073.1 KQ971354 EFA07014.1 BT127487 AEE62449.1 GEZM01057952 JAV72072.1 GEDC01026325 GEDC01011050 GEDC01010950 GEDC01002195 JAS10973.1 JAS26248.1 JAS26348.1 JAS35103.1 KQ414627 KOC67395.1 KK111565 KK107361 QOIP01000005 EZA46355.1 EZA52001.1 RLU22940.1 GL442767 EFN63078.1 KZ288230 PBC31574.1 JR051050 AEY61413.1 GL888147 EGI66562.1 KQ981724 KYN37243.1 ADTU01014477 ADTU01014478 LBMM01001284 KMQ96580.1 GEBQ01014756 JAT25221.1 GECU01029169 GECU01019377 JAS78537.1 JAS88329.1 GECZ01018001 JAS51768.1 NNAY01001451 OXU23937.1 GDRN01052400 JAI66306.1 DS235006 EEB10371.1 GECL01002810 JAP03314.1 GBBI01000550 JAC18162.1 GBGD01002956 JAC85933.1 GDKW01002354 JAI54241.1 ACPB03000375 GAHY01002009 JAA75501.1 GFTR01003447 JAW12979.1 AK417282 BAN20497.1 GEMB01006477 JAR96866.1 HACG01037910 CEK84775.1 GFDF01008325 JAV05759.1 AAPE02039917 GFDF01008390 JAV05694.1 GDIP01082213 JAM21502.1 RQTK01000184 RUS85043.1 GDIQ01253615 JAJ98109.1 HACA01021310 CDW38671.1 GDIP01180692 JAJ42710.1 GDIP01223597 JAI99804.1 GDIP01233500 JAI89901.1 GDIQ01190980 JAK60745.1 GDIP01182399 JAJ41003.1 HACA01021309 CDW38670.1 CVRI01000059 CRL03379.1
NWSH01000016 PCG80812.1 AK405074 BAM20476.1 AK401888 BAM18510.1 AGBW02012379 OWR45151.1 KQ459053 KPJ03979.1 GAIX01006192 JAA86368.1 BABH01018125 ODYU01002448 SOQ39945.1 GEZM01057951 JAV72073.1 KQ971354 EFA07014.1 BT127487 AEE62449.1 GEZM01057952 JAV72072.1 GEDC01026325 GEDC01011050 GEDC01010950 GEDC01002195 JAS10973.1 JAS26248.1 JAS26348.1 JAS35103.1 KQ414627 KOC67395.1 KK111565 KK107361 QOIP01000005 EZA46355.1 EZA52001.1 RLU22940.1 GL442767 EFN63078.1 KZ288230 PBC31574.1 JR051050 AEY61413.1 GL888147 EGI66562.1 KQ981724 KYN37243.1 ADTU01014477 ADTU01014478 LBMM01001284 KMQ96580.1 GEBQ01014756 JAT25221.1 GECU01029169 GECU01019377 JAS78537.1 JAS88329.1 GECZ01018001 JAS51768.1 NNAY01001451 OXU23937.1 GDRN01052400 JAI66306.1 DS235006 EEB10371.1 GECL01002810 JAP03314.1 GBBI01000550 JAC18162.1 GBGD01002956 JAC85933.1 GDKW01002354 JAI54241.1 ACPB03000375 GAHY01002009 JAA75501.1 GFTR01003447 JAW12979.1 AK417282 BAN20497.1 GEMB01006477 JAR96866.1 HACG01037910 CEK84775.1 GFDF01008325 JAV05759.1 AAPE02039917 GFDF01008390 JAV05694.1 GDIP01082213 JAM21502.1 RQTK01000184 RUS85043.1 GDIQ01253615 JAJ98109.1 HACA01021310 CDW38671.1 GDIP01180692 JAJ42710.1 GDIP01223597 JAI99804.1 GDIP01233500 JAI89901.1 GDIQ01190980 JAK60745.1 GDIP01182399 JAJ41003.1 HACA01021309 CDW38670.1 CVRI01000059 CRL03379.1
Proteomes
UP000053240
UP000283053
UP000218220
UP000007151
UP000053268
UP000005204
+ More
UP000192223 UP000007266 UP000053825 UP000053097 UP000279307 UP000000311 UP000242457 UP000005203 UP000007755 UP000078541 UP000005205 UP000036403 UP000215335 UP000009046 UP000079169 UP000015103 UP000001074 UP000271974 UP000183832
UP000192223 UP000007266 UP000053825 UP000053097 UP000279307 UP000000311 UP000242457 UP000005203 UP000007755 UP000078541 UP000005205 UP000036403 UP000215335 UP000009046 UP000079169 UP000015103 UP000001074 UP000271974 UP000183832
Pfam
PF00076 RRM_1
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
ProteinModelPortal
Q0ZAL2
A0A0N0PC56
A0A3S2LIF7
A0A2A4KAX8
I4DRD6
I4DKS0
+ More
A0A212EUL3 A0A194QEY6 S4PB04 H9IXG7 A0A2H1VGH8 A0A1W4WYD2 A0A1Y1LEM5 D6WR37 J3JW69 A0A1Y1LEJ1 A0A1B6EB13 A0A0L7R948 A0A026W8N4 E2AU35 A0A2A3EJC1 V9IJU3 A0A087ZZC6 F4WGU3 A0A151JUW6 A0A158NFG4 A0A0J7NW30 A0A1B6LN98 A0A1B6HV38 A0A1B6FNI8 A0A232F094 A0A0P4WCG7 E0VAG5 A0A3Q0IR86 A0A0V0G5F6 A0A023F9K9 A0A069DPF5 A0A0P4VTH3 R4FLS7 A0A224XY24 R4WRH0 A0A170VQN7 A0A0B7AXY8 A0A1L8DH27 G1P0Z9 A0A1L8DH72 A0A0P5WSI9 A0A3S1BJ78 A0A0P5FLT9 A0A0K2ULU7 A0A0P5BTJ8 A0A0P4YRM3 A0A0P4Y821 A0A0P5K4N8 A0A0P5BMM0 A0A0K2UKQ9 A0A1J1IT08
A0A212EUL3 A0A194QEY6 S4PB04 H9IXG7 A0A2H1VGH8 A0A1W4WYD2 A0A1Y1LEM5 D6WR37 J3JW69 A0A1Y1LEJ1 A0A1B6EB13 A0A0L7R948 A0A026W8N4 E2AU35 A0A2A3EJC1 V9IJU3 A0A087ZZC6 F4WGU3 A0A151JUW6 A0A158NFG4 A0A0J7NW30 A0A1B6LN98 A0A1B6HV38 A0A1B6FNI8 A0A232F094 A0A0P4WCG7 E0VAG5 A0A3Q0IR86 A0A0V0G5F6 A0A023F9K9 A0A069DPF5 A0A0P4VTH3 R4FLS7 A0A224XY24 R4WRH0 A0A170VQN7 A0A0B7AXY8 A0A1L8DH27 G1P0Z9 A0A1L8DH72 A0A0P5WSI9 A0A3S1BJ78 A0A0P5FLT9 A0A0K2ULU7 A0A0P5BTJ8 A0A0P4YRM3 A0A0P4Y821 A0A0P5K4N8 A0A0P5BMM0 A0A0K2UKQ9 A0A1J1IT08
PDB
3B4M
E-value=7.48984e-40,
Score=408
Ontologies
KEGG
GO
PANTHER
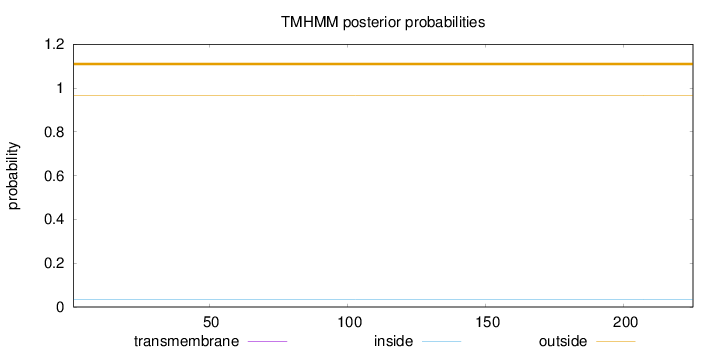
Topology
Length:
225
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00134
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03540
outside
1 - 225
Population Genetic Test Statistics
Pi
153.291223
Theta
143.34003
Tajima's D
0.297537
CLR
0.795967
CSRT
0.454377281135943
Interpretation
Uncertain
