Pre Gene Modal
BGIBMGA001868
Annotation
PREDICTED:_vacuolar_membrane_protein_YPL162C-like_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.966
Sequence
CDS
ATGAATAATAGTTCGGATTACATATACTGGTCCGAGCCGCATTGCTCGAAGGATGCTCTTACCGATACATATGGGTGGTTCATGCAAATTATTTTGGCAGTTTTAGCATTTACCTGTTTAATAGGTAAGAGATTTTGTGAACCACGTTTCGCCAGAAGGCCATGGCTGATATGGTTTTACGACACATCAAAGCAAGGCTTAGGAGCTCTTATAATTCATGCAGCAAATGTTTGGCTCTCACCACATCTTACCGGAAACCCATGCACATGGTACATTGTAAACTTTATGTTGGATTCAACTTTAGGTTTATTGATAATATGGGCGGGAATCAGATTAGCTCAGCACTGTGCAAGAGTTTATGACATACCATTGATAAATTTTGGTGAATATGGTAAACCTCCAATGTGCTCCGCGTGGATATGCCAGTGTATATTATACGCGGCTCTAGCAACATTTGCCAAGTCTGTCCTGGCGCTGGTGTTGCGCCTCCCGCCTGTGGTAGCGGTGCTGTCCACGTTGAGACTGTCTCCGGTGTCCGATCCGCGCCTGGAGCTAGCTGTCGTTATGCTTATAATACCATTCTTTGTTAATATACTAATATTCTGGGTGACAGATAACTTCCTAATGTATCATCCCAGAGGAGTCAGTACTAAAATTAAAACTAAGGTGAGATACCAATCGATAAAGAAAGAGAAGTCTGGTTCGGAAGAGGACGAGCACTCGGCCGACGAGCGGCTGCTCGGCGCTAGCGTATGA
Protein
MNNSSDYIYWSEPHCSKDALTDTYGWFMQIILAVLAFTCLIGKRFCEPRFARRPWLIWFYDTSKQGLGALIIHAANVWLSPHLTGNPCTWYIVNFMLDSTLGLLIIWAGIRLAQHCARVYDIPLINFGEYGKPPMCSAWICQCILYAALATFAKSVLALVLRLPPVVAVLSTLRLSPVSDPRLELAVVMLIIPFFVNILIFWVTDNFLMYHPRGVSTKIKTKVRYQSIKKEKSGSEEDEHSADERLLGASV
Summary
Cofactor
NAD(+)
Similarity
Belongs to the NAD(P)-dependent epimerase/dehydratase family.
Uniprot
H9IX85
A0A2H1VHQ6
A0A0N1PGN7
A0A2A4KAU8
A0A194QKK9
A0A3S2NCS2
+ More
A0A212EM88 A0A0L7LDQ2 A0A0L7L6G3 J3JXR3 D6WR74 U4UMK0 A0A067QJ61 N6TRZ7 N6SW95 N6T394 A0A1B6C1Z1 A0A1Y1JWX3 Q173W9 B0W2Y0 A0A0K8TQ15 A0A1Q3F747 T1DGF2 A0NE32 A0A182YM81 A0A182M7H8 A0A182IBH3 A0A182JSV1 A0A182WFD8 A0A1S4GYW1 A0A182TUJ2 A0A182X4Y1 A0A182VN47 A0A182PJF4 U5ESR1 A0A1L8D9A8 A0A182QTW4 A0A2M3Z3C3 Q173W8 A0A0P6JSP6 A0A182H8E7 A0A1Y9HFA1 A0A2J7R5J1 A0A182KN28 A0A182J4F5 A0A1B0D4R1 A0A2P8YYM5 A0A182NGS1 E0VSA6 A0A1B6HKD3 A0A2M4BXD5 A0A2M3Z6S8 A0A1B6H4E6 A0A2M4AJC3 T1DD43 W5JMK9 A0A293MCI1 A0A084VJ67 A0A0N7ZDR7 A0A182F383 A0A2H8TYS4 J9JQZ6 A0A2S2P1Y0 E9GKQ7 T1D2P0 A0A0N8BCC9 A0A0P6HT51 A0A0P5X000 J9JYF6 A0A0P4ZPW0 A0A444TB04 A0A2S2PX34 K1QSV3 A0A1J1IQG9 A0A2T7NT49 A0A433TS84 A0A210Q4G9 F1NIC7 R7UMZ6 A0A2R5L9J6 A0A3B3Z9X1 A0A437DDQ6 A0A3B3QX01 A0A3B3QXJ9 C3YAD5 A0A3P9KP23 A0A218VCL7 A0A452HN41 H2LUG1 A0A3P9HDV0 A0A0P4XSS1 A0A2H6N078 A0A3P8V0Y1 A0A1A8K918 A0A1A8BG99 A0A1A8AVZ4 A0A0F8ASB8 A0A1A8J021
A0A212EM88 A0A0L7LDQ2 A0A0L7L6G3 J3JXR3 D6WR74 U4UMK0 A0A067QJ61 N6TRZ7 N6SW95 N6T394 A0A1B6C1Z1 A0A1Y1JWX3 Q173W9 B0W2Y0 A0A0K8TQ15 A0A1Q3F747 T1DGF2 A0NE32 A0A182YM81 A0A182M7H8 A0A182IBH3 A0A182JSV1 A0A182WFD8 A0A1S4GYW1 A0A182TUJ2 A0A182X4Y1 A0A182VN47 A0A182PJF4 U5ESR1 A0A1L8D9A8 A0A182QTW4 A0A2M3Z3C3 Q173W8 A0A0P6JSP6 A0A182H8E7 A0A1Y9HFA1 A0A2J7R5J1 A0A182KN28 A0A182J4F5 A0A1B0D4R1 A0A2P8YYM5 A0A182NGS1 E0VSA6 A0A1B6HKD3 A0A2M4BXD5 A0A2M3Z6S8 A0A1B6H4E6 A0A2M4AJC3 T1DD43 W5JMK9 A0A293MCI1 A0A084VJ67 A0A0N7ZDR7 A0A182F383 A0A2H8TYS4 J9JQZ6 A0A2S2P1Y0 E9GKQ7 T1D2P0 A0A0N8BCC9 A0A0P6HT51 A0A0P5X000 J9JYF6 A0A0P4ZPW0 A0A444TB04 A0A2S2PX34 K1QSV3 A0A1J1IQG9 A0A2T7NT49 A0A433TS84 A0A210Q4G9 F1NIC7 R7UMZ6 A0A2R5L9J6 A0A3B3Z9X1 A0A437DDQ6 A0A3B3QX01 A0A3B3QXJ9 C3YAD5 A0A3P9KP23 A0A218VCL7 A0A452HN41 H2LUG1 A0A3P9HDV0 A0A0P4XSS1 A0A2H6N078 A0A3P8V0Y1 A0A1A8K918 A0A1A8BG99 A0A1A8AVZ4 A0A0F8ASB8 A0A1A8J021
Pubmed
19121390
26354079
22118469
26227816
22516182
18362917
+ More
19820115 23537049 24845553 28004739 17510324 26369729 24330624 12364791 25244985 26999592 26483478 20966253 29403074 20566863 20920257 23761445 24438588 21292972 22992520 28812685 15592404 23254933 25463417 29240929 18563158 17554307 28562605 24487278 25835551
19820115 23537049 24845553 28004739 17510324 26369729 24330624 12364791 25244985 26999592 26483478 20966253 29403074 20566863 20920257 23761445 24438588 21292972 22992520 28812685 15592404 23254933 25463417 29240929 18563158 17554307 28562605 24487278 25835551
EMBL
BABH01018130
ODYU01002448
SOQ39952.1
KQ460772
KPJ12458.1
NWSH01000016
+ More
PCG80782.1 KQ459053 KPJ03986.1 RSAL01000096 RVE47758.1 AGBW02013910 OWR42571.1 JTDY01001624 KOB73316.1 JTDY01002693 KOB70904.1 BT128034 AEE62995.1 KQ971354 EFA05999.1 KB632281 ERL91366.1 KK853284 KDR08976.1 APGK01054138 APGK01054139 KB741247 ENN72030.1 ENN72029.1 ENN72028.1 GEDC01030038 GEDC01018001 GEDC01017561 GEDC01016479 GEDC01014915 JAS07260.1 JAS19297.1 JAS19737.1 JAS20819.1 JAS22383.1 GEZM01101805 JAV52290.1 CH477416 EAT41327.1 DS231829 EDS30339.1 GDAI01001121 JAI16482.1 GFDL01011659 JAV23386.1 GALA01000202 JAA94650.1 AAAB01008944 EAU76704.2 AXCM01009206 APCN01000690 GANO01003120 JAB56751.1 GFDF01011043 JAV03041.1 AXCN02002201 GGFM01002261 MBW23012.1 EAT41328.1 GDUN01000193 JAN95726.1 JXUM01029822 JXUM01029823 JXUM01029824 KQ560865 KXJ80574.1 NEVH01006994 PNF36105.1 AJVK01011578 PYGN01000284 PSN49354.1 DS235748 EEB16262.1 GECU01032564 GECU01018971 GECU01000482 JAS75142.1 JAS88735.1 JAT07225.1 GGFJ01008595 MBW57736.1 GGFM01003476 MBW24227.1 GECZ01000295 JAS69474.1 GGFK01007564 MBW40885.1 GALA01001562 JAA93290.1 ADMH02001090 ETN64140.1 GFWV01012969 MAA37698.1 ATLV01013473 ATLV01013474 KE524860 KFB38011.1 GDRN01002376 JAI68000.1 GFXV01007235 MBW19040.1 ABLF02008307 GGMR01010765 MBY23384.1 GL732550 EFX79927.1 GALA01001553 JAA93299.1 GDIQ01191627 JAK60098.1 GDIQ01015314 JAN79423.1 GDIP01091309 LRGB01003257 JAM12406.1 KZS03563.1 ABLF02022728 GDIP01222899 JAJ00503.1 SAUD01005606 RXG67152.1 GGMS01000717 MBY69920.1 JH817059 EKC24671.1 CVRI01000056 CRL01788.1 PZQS01000009 PVD24334.1 RQTK01000206 RUS84434.1 NEDP02005055 OWF43579.1 AADN05000356 AMQN01008082 KB302153 ELU04766.1 GGLE01002050 MBY06176.1 CM012441 RVE72894.1 GG666494 EEN62683.1 MUZQ01000011 OWK63588.1 GDIP01238251 JAI85150.1 IACI01022423 LAA21704.1 HAEE01008870 SBR28920.1 HADZ01001725 SBP65666.1 HADY01020714 HAEJ01014051 SBP59199.1 KQ042855 KKF09404.1 HAED01016535 SBR02980.1
PCG80782.1 KQ459053 KPJ03986.1 RSAL01000096 RVE47758.1 AGBW02013910 OWR42571.1 JTDY01001624 KOB73316.1 JTDY01002693 KOB70904.1 BT128034 AEE62995.1 KQ971354 EFA05999.1 KB632281 ERL91366.1 KK853284 KDR08976.1 APGK01054138 APGK01054139 KB741247 ENN72030.1 ENN72029.1 ENN72028.1 GEDC01030038 GEDC01018001 GEDC01017561 GEDC01016479 GEDC01014915 JAS07260.1 JAS19297.1 JAS19737.1 JAS20819.1 JAS22383.1 GEZM01101805 JAV52290.1 CH477416 EAT41327.1 DS231829 EDS30339.1 GDAI01001121 JAI16482.1 GFDL01011659 JAV23386.1 GALA01000202 JAA94650.1 AAAB01008944 EAU76704.2 AXCM01009206 APCN01000690 GANO01003120 JAB56751.1 GFDF01011043 JAV03041.1 AXCN02002201 GGFM01002261 MBW23012.1 EAT41328.1 GDUN01000193 JAN95726.1 JXUM01029822 JXUM01029823 JXUM01029824 KQ560865 KXJ80574.1 NEVH01006994 PNF36105.1 AJVK01011578 PYGN01000284 PSN49354.1 DS235748 EEB16262.1 GECU01032564 GECU01018971 GECU01000482 JAS75142.1 JAS88735.1 JAT07225.1 GGFJ01008595 MBW57736.1 GGFM01003476 MBW24227.1 GECZ01000295 JAS69474.1 GGFK01007564 MBW40885.1 GALA01001562 JAA93290.1 ADMH02001090 ETN64140.1 GFWV01012969 MAA37698.1 ATLV01013473 ATLV01013474 KE524860 KFB38011.1 GDRN01002376 JAI68000.1 GFXV01007235 MBW19040.1 ABLF02008307 GGMR01010765 MBY23384.1 GL732550 EFX79927.1 GALA01001553 JAA93299.1 GDIQ01191627 JAK60098.1 GDIQ01015314 JAN79423.1 GDIP01091309 LRGB01003257 JAM12406.1 KZS03563.1 ABLF02022728 GDIP01222899 JAJ00503.1 SAUD01005606 RXG67152.1 GGMS01000717 MBY69920.1 JH817059 EKC24671.1 CVRI01000056 CRL01788.1 PZQS01000009 PVD24334.1 RQTK01000206 RUS84434.1 NEDP02005055 OWF43579.1 AADN05000356 AMQN01008082 KB302153 ELU04766.1 GGLE01002050 MBY06176.1 CM012441 RVE72894.1 GG666494 EEN62683.1 MUZQ01000011 OWK63588.1 GDIP01238251 JAI85150.1 IACI01022423 LAA21704.1 HAEE01008870 SBR28920.1 HADZ01001725 SBP65666.1 HADY01020714 HAEJ01014051 SBP59199.1 KQ042855 KKF09404.1 HAED01016535 SBR02980.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000283053
UP000007151
+ More
UP000037510 UP000007266 UP000030742 UP000027135 UP000019118 UP000008820 UP000002320 UP000007062 UP000076408 UP000075883 UP000075840 UP000075881 UP000075920 UP000075902 UP000076407 UP000075903 UP000075885 UP000075886 UP000069940 UP000249989 UP000075900 UP000235965 UP000075882 UP000075880 UP000092462 UP000245037 UP000075884 UP000009046 UP000000673 UP000030765 UP000069272 UP000007819 UP000000305 UP000076858 UP000288706 UP000005408 UP000183832 UP000245119 UP000271974 UP000242188 UP000000539 UP000014760 UP000261520 UP000261540 UP000001554 UP000265180 UP000197619 UP000291020 UP000001038 UP000265200 UP000265120
UP000037510 UP000007266 UP000030742 UP000027135 UP000019118 UP000008820 UP000002320 UP000007062 UP000076408 UP000075883 UP000075840 UP000075881 UP000075920 UP000075902 UP000076407 UP000075903 UP000075885 UP000075886 UP000069940 UP000249989 UP000075900 UP000235965 UP000075882 UP000075880 UP000092462 UP000245037 UP000075884 UP000009046 UP000000673 UP000030765 UP000069272 UP000007819 UP000000305 UP000076858 UP000288706 UP000005408 UP000183832 UP000245119 UP000271974 UP000242188 UP000000539 UP000014760 UP000261520 UP000261540 UP000001554 UP000265180 UP000197619 UP000291020 UP000001038 UP000265200 UP000265120
PRIDE
Pfam
Interpro
IPR022127
STIMATE/YPL162C
+ More
IPR020846 MFS_dom
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR016187 CTDL_fold
IPR005886 UDP_G4E
IPR036291 NAD(P)-bd_dom_sf
IPR016040 NAD(P)-bd_dom
IPR007245 PIG-T
IPR011322 N-reg_PII-like_a/b
IPR004323 Ion_tolerance_CutA
IPR015867 N-reg_PII/ATP_PRibTrfase_C
IPR020846 MFS_dom
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR016187 CTDL_fold
IPR005886 UDP_G4E
IPR036291 NAD(P)-bd_dom_sf
IPR016040 NAD(P)-bd_dom
IPR007245 PIG-T
IPR011322 N-reg_PII-like_a/b
IPR004323 Ion_tolerance_CutA
IPR015867 N-reg_PII/ATP_PRibTrfase_C
Gene 3D
ProteinModelPortal
H9IX85
A0A2H1VHQ6
A0A0N1PGN7
A0A2A4KAU8
A0A194QKK9
A0A3S2NCS2
+ More
A0A212EM88 A0A0L7LDQ2 A0A0L7L6G3 J3JXR3 D6WR74 U4UMK0 A0A067QJ61 N6TRZ7 N6SW95 N6T394 A0A1B6C1Z1 A0A1Y1JWX3 Q173W9 B0W2Y0 A0A0K8TQ15 A0A1Q3F747 T1DGF2 A0NE32 A0A182YM81 A0A182M7H8 A0A182IBH3 A0A182JSV1 A0A182WFD8 A0A1S4GYW1 A0A182TUJ2 A0A182X4Y1 A0A182VN47 A0A182PJF4 U5ESR1 A0A1L8D9A8 A0A182QTW4 A0A2M3Z3C3 Q173W8 A0A0P6JSP6 A0A182H8E7 A0A1Y9HFA1 A0A2J7R5J1 A0A182KN28 A0A182J4F5 A0A1B0D4R1 A0A2P8YYM5 A0A182NGS1 E0VSA6 A0A1B6HKD3 A0A2M4BXD5 A0A2M3Z6S8 A0A1B6H4E6 A0A2M4AJC3 T1DD43 W5JMK9 A0A293MCI1 A0A084VJ67 A0A0N7ZDR7 A0A182F383 A0A2H8TYS4 J9JQZ6 A0A2S2P1Y0 E9GKQ7 T1D2P0 A0A0N8BCC9 A0A0P6HT51 A0A0P5X000 J9JYF6 A0A0P4ZPW0 A0A444TB04 A0A2S2PX34 K1QSV3 A0A1J1IQG9 A0A2T7NT49 A0A433TS84 A0A210Q4G9 F1NIC7 R7UMZ6 A0A2R5L9J6 A0A3B3Z9X1 A0A437DDQ6 A0A3B3QX01 A0A3B3QXJ9 C3YAD5 A0A3P9KP23 A0A218VCL7 A0A452HN41 H2LUG1 A0A3P9HDV0 A0A0P4XSS1 A0A2H6N078 A0A3P8V0Y1 A0A1A8K918 A0A1A8BG99 A0A1A8AVZ4 A0A0F8ASB8 A0A1A8J021
A0A212EM88 A0A0L7LDQ2 A0A0L7L6G3 J3JXR3 D6WR74 U4UMK0 A0A067QJ61 N6TRZ7 N6SW95 N6T394 A0A1B6C1Z1 A0A1Y1JWX3 Q173W9 B0W2Y0 A0A0K8TQ15 A0A1Q3F747 T1DGF2 A0NE32 A0A182YM81 A0A182M7H8 A0A182IBH3 A0A182JSV1 A0A182WFD8 A0A1S4GYW1 A0A182TUJ2 A0A182X4Y1 A0A182VN47 A0A182PJF4 U5ESR1 A0A1L8D9A8 A0A182QTW4 A0A2M3Z3C3 Q173W8 A0A0P6JSP6 A0A182H8E7 A0A1Y9HFA1 A0A2J7R5J1 A0A182KN28 A0A182J4F5 A0A1B0D4R1 A0A2P8YYM5 A0A182NGS1 E0VSA6 A0A1B6HKD3 A0A2M4BXD5 A0A2M3Z6S8 A0A1B6H4E6 A0A2M4AJC3 T1DD43 W5JMK9 A0A293MCI1 A0A084VJ67 A0A0N7ZDR7 A0A182F383 A0A2H8TYS4 J9JQZ6 A0A2S2P1Y0 E9GKQ7 T1D2P0 A0A0N8BCC9 A0A0P6HT51 A0A0P5X000 J9JYF6 A0A0P4ZPW0 A0A444TB04 A0A2S2PX34 K1QSV3 A0A1J1IQG9 A0A2T7NT49 A0A433TS84 A0A210Q4G9 F1NIC7 R7UMZ6 A0A2R5L9J6 A0A3B3Z9X1 A0A437DDQ6 A0A3B3QX01 A0A3B3QXJ9 C3YAD5 A0A3P9KP23 A0A218VCL7 A0A452HN41 H2LUG1 A0A3P9HDV0 A0A0P4XSS1 A0A2H6N078 A0A3P8V0Y1 A0A1A8K918 A0A1A8BG99 A0A1A8AVZ4 A0A0F8ASB8 A0A1A8J021
Ontologies
GO
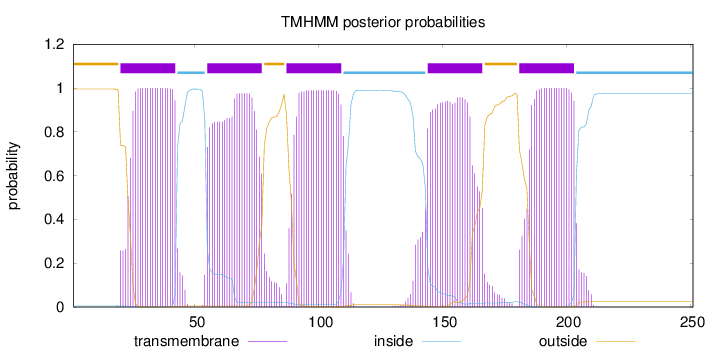
Topology
Length:
251
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
106.64493
Exp number, first 60 AAs:
25.52349
Total prob of N-in:
0.00492
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 54
TMhelix
55 - 77
outside
78 - 86
TMhelix
87 - 109
inside
110 - 143
TMhelix
144 - 166
outside
167 - 180
TMhelix
181 - 203
inside
204 - 251
Population Genetic Test Statistics
Pi
275.664704
Theta
204.67511
Tajima's D
0.603387
CLR
0.043484
CSRT
0.541572921353932
Interpretation
Uncertain
