Gene
KWMTBOMO11389
Pre Gene Modal
BGIBMGA001864
Annotation
PREDICTED:_trimeric_intracellular_cation_channel_type_B_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 3.905
Sequence
CDS
ATGGATCCTGAATCATTTTTAGACCTGGCCAATCAGGTCATAAAGTTGAAAATGTATCCATATTTCGACATAGCGCATTCGTTGCTGTGCGCTCTTGCAGTCAGAGAAGATTTGGGATCCGGTGCCCAGGCCTTTTCCAGGAAGCATCCACTGTCATGCTGGCTGTCCACTATGCTAGTGATATTCGCCGGCGGTATGGTAGCCAATGGTCTTCTAGGCGAACCCATTTTGGCGCCGTTGAAGAATACACCCCAATTAGTCATCGGAACTGTTACATGGTACTTAGTTTTCTACACGCCTTTCGATGTAGGCTACAAGGTAGCGAAGTTCCTACCGGTGAAGGTGGTGGCGGCTGCGATGAAAGAAATATACAGAGCCAAGAAAGTTTACGATGGAGTCAGCCACGCAGCCAAACTGTACCCGAACGCTTACGTCATTATGGTCATTGTTGGTACGTTGAAAGGTAACGGTGCCGGCTTCACGAAATTAGTCGAGAGGCTTATCCGGGGCGCTTGGACCCCGACTGCGATGGAAACCATGCAACCCAGCTTCTACACAAAGGCTTCGCTGGTAGCTTCCGTGATCTTCGTATTGGATAAGAAGACAGATTTGATCTCTGCTCCCCACGCACTCGTCTACTTCGGTATCGTTATCTTCTTTGTTTACTTTAAGCTCTCCTCCATACTTCTGGGCATCCACGATCCCTTCGTGCCATTTGAGAATCTGTTCTGCGCCTTATTCATGGGCGGCATTTGGGATTCGCTGGCCAAGTTGCTCGGCAAAGGACAACCCAAGGAGGAGACTAAGGACACCAAGAAAATCAACTAG
Protein
MDPESFLDLANQVIKLKMYPYFDIAHSLLCALAVREDLGSGAQAFSRKHPLSCWLSTMLVIFAGGMVANGLLGEPILAPLKNTPQLVIGTVTWYLVFYTPFDVGYKVAKFLPVKVVAAAMKEIYRAKKVYDGVSHAAKLYPNAYVIMVIVGTLKGNGAGFTKLVERLIRGAWTPTAMETMQPSFYTKASLVASVIFVLDKKTDLISAPHALVYFGIVIFFVYFKLSSILLGIHDPFVPFENLFCALFMGGIWDSLAKLLGKGQPKEETKDTKKIN
Summary
Similarity
Belongs to the synaptobrevin family.
Uniprot
A0A2A4KA55
A0A194QF01
A0A2H1WNH3
I4DN83
A0A0N0PBX8
A0A1E1WN10
+ More
A0A437BBF2 A0A1B0CWA4 A0A1L8DDN2 B0WYR2 Q1HRE2 A0A182G698 A0A1Q3G471 U5ESJ9 T1DN85 A0A2M3ZB48 A0A2M4BWT2 W5JKT8 A0A182FJK1 A0A2M4AX15 A0A084VVH8 A0A182JH44 A0A182YH80 A0A182PSV7 A0A182LU20 A0A182SYP8 A0A1Y9HB63 A0A182KD93 A0A182U8F4 A0A182W5N2 A0A182XJX9 A0NCG5 A0A182RHL7 A0A182IAV9 W8CA05 A0A0L0C3W2 A0A034WCS9 A0A2Y9D318 A0A0K8VWZ5 A0A034WA63 A0A182N8G9 T1PBK4 A0A023ELS4 A0A1I8PT06 B3N0S0 B4M1S4 A0A026WTP1 B4R5T6 B4IF71 Q9VXG9 B4PXS7 B4L8R6 A0A1W4VHD7 A0A3B0KQ72 B4H0Z9 A0A0M5J5W1 Q29GI6 B4JKM4 B3NX45 E2BL30 D6WU90 A0A232FGQ6 E2AIS7 B4NEV3 E0W2J7 E9J0B0 V5G537 J3JWM4 A0A0L7RC96 A0A1B6K1J0 A0A2A3E5L9 A0A087ZY47 A0A1Y1MJQ4 A0A154P8Q8 A0A158NPX4 A0A1A9VRF7 A0A336MRA2 A0A067QTZ9 A0A1A9XDX3 A0A151IVK9 A0A195BET1 A0A195FH63 A0A1B0DNP4 A0A151IDD0 A0A1B6F8I2 A0A0N0BE69 A0A2J7R9A1 A0A1B6L944 A0A1S3DH01 A0A0A9XH50 A0A0P4VY36 R4WIN7 A0A224XVJ8 A0A0V0GC56 A0A069DR86 A0A1Q3G4B7 A0A0C9S187 T1HGV4 A0A1B6DH47 A0A182KJH1
A0A437BBF2 A0A1B0CWA4 A0A1L8DDN2 B0WYR2 Q1HRE2 A0A182G698 A0A1Q3G471 U5ESJ9 T1DN85 A0A2M3ZB48 A0A2M4BWT2 W5JKT8 A0A182FJK1 A0A2M4AX15 A0A084VVH8 A0A182JH44 A0A182YH80 A0A182PSV7 A0A182LU20 A0A182SYP8 A0A1Y9HB63 A0A182KD93 A0A182U8F4 A0A182W5N2 A0A182XJX9 A0NCG5 A0A182RHL7 A0A182IAV9 W8CA05 A0A0L0C3W2 A0A034WCS9 A0A2Y9D318 A0A0K8VWZ5 A0A034WA63 A0A182N8G9 T1PBK4 A0A023ELS4 A0A1I8PT06 B3N0S0 B4M1S4 A0A026WTP1 B4R5T6 B4IF71 Q9VXG9 B4PXS7 B4L8R6 A0A1W4VHD7 A0A3B0KQ72 B4H0Z9 A0A0M5J5W1 Q29GI6 B4JKM4 B3NX45 E2BL30 D6WU90 A0A232FGQ6 E2AIS7 B4NEV3 E0W2J7 E9J0B0 V5G537 J3JWM4 A0A0L7RC96 A0A1B6K1J0 A0A2A3E5L9 A0A087ZY47 A0A1Y1MJQ4 A0A154P8Q8 A0A158NPX4 A0A1A9VRF7 A0A336MRA2 A0A067QTZ9 A0A1A9XDX3 A0A151IVK9 A0A195BET1 A0A195FH63 A0A1B0DNP4 A0A151IDD0 A0A1B6F8I2 A0A0N0BE69 A0A2J7R9A1 A0A1B6L944 A0A1S3DH01 A0A0A9XH50 A0A0P4VY36 R4WIN7 A0A224XVJ8 A0A0V0GC56 A0A069DR86 A0A1Q3G4B7 A0A0C9S187 T1HGV4 A0A1B6DH47 A0A182KJH1
Pubmed
26354079
22651552
17204158
17510324
26483478
20920257
+ More
23761445 24438588 25244985 12364791 14747013 17210077 24495485 26108605 25348373 25315136 24945155 17994087 18057021 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 23185243 20798317 18362917 19820115 28648823 20566863 21282665 22516182 23537049 28004739 21347285 24845553 25401762 26823975 27129103 23691247 26334808 20966253
23761445 24438588 25244985 12364791 14747013 17210077 24495485 26108605 25348373 25315136 24945155 17994087 18057021 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 23185243 20798317 18362917 19820115 28648823 20566863 21282665 22516182 23537049 28004739 21347285 24845553 25401762 26823975 27129103 23691247 26334808 20966253
EMBL
NWSH01000016
PCG80794.1
KQ459053
KPJ03994.1
ODYU01009504
SOQ53994.1
+ More
AK402768 BAM19373.1 KQ460772 KPJ12451.1 GDQN01002680 JAT88374.1 RSAL01000096 RVE47750.1 AJWK01032126 GFDF01009536 JAV04548.1 DS232196 DS232309 EDS37172.1 EDS39565.1 DQ440152 CH477242 ABF18185.1 EAT46348.1 EAT46349.1 EAT46350.1 JXUM01007375 JXUM01007376 JXUM01007377 JXUM01007378 JXUM01044523 KQ561404 KXJ78691.1 GFDL01000440 JAV34605.1 GANO01003207 JAB56664.1 GAMD01003105 JAA98485.1 GGFM01004982 MBW25733.1 GGFJ01008250 MBW57391.1 ADMH02000803 GGFL01004300 ETN64982.1 MBW68478.1 GGFK01011931 MBW45252.1 ATLV01017184 KE525157 KFB41972.1 AXCP01004575 AXCM01004821 AXCN02002233 AAAB01008846 EAA45107.1 EGK96463.1 APCN01001348 GAMC01001694 JAC04862.1 JRES01000948 KNC26901.1 GAKP01007364 JAC51588.1 GDHF01008917 JAI43397.1 GAKP01007363 JAC51589.1 KA645353 AFP59982.1 GAPW01003380 JAC10218.1 CH902644 EDV34799.1 KPU75287.1 CH940651 EDW65628.1 KRF82314.1 KRF82315.1 KK107106 QOIP01000002 EZA59430.1 RLU25732.1 CM000366 EDX18086.1 CH480832 EDW46125.1 AE014298 AY060786 AAF48602.1 AAL28334.1 CM000162 EDX01913.1 KRK06439.1 CH933815 EDW08041.1 KRG07571.1 KRG07572.1 KRG07573.1 OUUW01000015 SPP88769.1 CH479201 EDW29915.1 CP012528 ALC48586.1 CH379064 EAL32123.1 KRT06947.1 KRT06948.1 KRT06949.1 CH916370 EDW00127.1 CH954180 EDV46874.1 KQS30218.1 KQS30219.1 KQS30220.1 GL448906 EFN83599.1 KQ971352 EFA06252.1 NNAY01000245 OXU29743.1 GL439874 EFN66666.1 CH964239 EDW82272.1 KRF99589.1 DS235878 EEB19853.1 GL767381 EFZ13739.1 GALX01003291 JAB65175.1 APGK01057806 BT127642 KB741282 KB631802 AEE62604.1 ENN70797.1 ERL86249.1 KQ414616 KOC68453.1 GECU01002401 JAT05306.1 KZ288369 PBC26774.1 GEZM01029765 JAV85941.1 KQ434829 KZC07744.1 ADTU01000325 ADTU01000326 ADTU01000327 UFQT01000175 UFQT01001969 SSX21146.1 SSX32235.1 KK852954 KDR13304.1 KQ980895 KYN11737.1 KQ976509 KYM82692.1 KQ981606 KYN39572.1 AJVK01007642 KQ977978 KYM98396.1 GECZ01023242 JAS46527.1 KQ435847 KOX71141.1 NEVH01006600 PNF37403.1 GEBQ01019812 JAT20165.1 GBHO01024285 GBRD01000271 GDHC01006090 JAG19319.1 JAG65550.1 JAQ12539.1 GDKW01001056 JAI55539.1 AK417315 BAN20530.1 GFTR01004407 JAW12019.1 GECL01000490 JAP05634.1 GBGD01002514 JAC86375.1 GFDL01000439 JAV34606.1 GBYB01014516 JAG84283.1 ACPB03000101 GEDC01012326 JAS24972.1
AK402768 BAM19373.1 KQ460772 KPJ12451.1 GDQN01002680 JAT88374.1 RSAL01000096 RVE47750.1 AJWK01032126 GFDF01009536 JAV04548.1 DS232196 DS232309 EDS37172.1 EDS39565.1 DQ440152 CH477242 ABF18185.1 EAT46348.1 EAT46349.1 EAT46350.1 JXUM01007375 JXUM01007376 JXUM01007377 JXUM01007378 JXUM01044523 KQ561404 KXJ78691.1 GFDL01000440 JAV34605.1 GANO01003207 JAB56664.1 GAMD01003105 JAA98485.1 GGFM01004982 MBW25733.1 GGFJ01008250 MBW57391.1 ADMH02000803 GGFL01004300 ETN64982.1 MBW68478.1 GGFK01011931 MBW45252.1 ATLV01017184 KE525157 KFB41972.1 AXCP01004575 AXCM01004821 AXCN02002233 AAAB01008846 EAA45107.1 EGK96463.1 APCN01001348 GAMC01001694 JAC04862.1 JRES01000948 KNC26901.1 GAKP01007364 JAC51588.1 GDHF01008917 JAI43397.1 GAKP01007363 JAC51589.1 KA645353 AFP59982.1 GAPW01003380 JAC10218.1 CH902644 EDV34799.1 KPU75287.1 CH940651 EDW65628.1 KRF82314.1 KRF82315.1 KK107106 QOIP01000002 EZA59430.1 RLU25732.1 CM000366 EDX18086.1 CH480832 EDW46125.1 AE014298 AY060786 AAF48602.1 AAL28334.1 CM000162 EDX01913.1 KRK06439.1 CH933815 EDW08041.1 KRG07571.1 KRG07572.1 KRG07573.1 OUUW01000015 SPP88769.1 CH479201 EDW29915.1 CP012528 ALC48586.1 CH379064 EAL32123.1 KRT06947.1 KRT06948.1 KRT06949.1 CH916370 EDW00127.1 CH954180 EDV46874.1 KQS30218.1 KQS30219.1 KQS30220.1 GL448906 EFN83599.1 KQ971352 EFA06252.1 NNAY01000245 OXU29743.1 GL439874 EFN66666.1 CH964239 EDW82272.1 KRF99589.1 DS235878 EEB19853.1 GL767381 EFZ13739.1 GALX01003291 JAB65175.1 APGK01057806 BT127642 KB741282 KB631802 AEE62604.1 ENN70797.1 ERL86249.1 KQ414616 KOC68453.1 GECU01002401 JAT05306.1 KZ288369 PBC26774.1 GEZM01029765 JAV85941.1 KQ434829 KZC07744.1 ADTU01000325 ADTU01000326 ADTU01000327 UFQT01000175 UFQT01001969 SSX21146.1 SSX32235.1 KK852954 KDR13304.1 KQ980895 KYN11737.1 KQ976509 KYM82692.1 KQ981606 KYN39572.1 AJVK01007642 KQ977978 KYM98396.1 GECZ01023242 JAS46527.1 KQ435847 KOX71141.1 NEVH01006600 PNF37403.1 GEBQ01019812 JAT20165.1 GBHO01024285 GBRD01000271 GDHC01006090 JAG19319.1 JAG65550.1 JAQ12539.1 GDKW01001056 JAI55539.1 AK417315 BAN20530.1 GFTR01004407 JAW12019.1 GECL01000490 JAP05634.1 GBGD01002514 JAC86375.1 GFDL01000439 JAV34606.1 GBYB01014516 JAG84283.1 ACPB03000101 GEDC01012326 JAS24972.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000092461
UP000002320
+ More
UP000008820 UP000069940 UP000249989 UP000000673 UP000069272 UP000030765 UP000075880 UP000076408 UP000075885 UP000075883 UP000075901 UP000075886 UP000075881 UP000075902 UP000075920 UP000076407 UP000007062 UP000075900 UP000075840 UP000037069 UP000075903 UP000075884 UP000095301 UP000095300 UP000007801 UP000008792 UP000053097 UP000279307 UP000000304 UP000001292 UP000000803 UP000002282 UP000009192 UP000192221 UP000268350 UP000008744 UP000092553 UP000001819 UP000001070 UP000008711 UP000008237 UP000007266 UP000215335 UP000000311 UP000007798 UP000009046 UP000019118 UP000030742 UP000053825 UP000242457 UP000005203 UP000076502 UP000005205 UP000078200 UP000027135 UP000092443 UP000078492 UP000078540 UP000078541 UP000092462 UP000078542 UP000053105 UP000235965 UP000079169 UP000015103 UP000075882
UP000008820 UP000069940 UP000249989 UP000000673 UP000069272 UP000030765 UP000075880 UP000076408 UP000075885 UP000075883 UP000075901 UP000075886 UP000075881 UP000075902 UP000075920 UP000076407 UP000007062 UP000075900 UP000075840 UP000037069 UP000075903 UP000075884 UP000095301 UP000095300 UP000007801 UP000008792 UP000053097 UP000279307 UP000000304 UP000001292 UP000000803 UP000002282 UP000009192 UP000192221 UP000268350 UP000008744 UP000092553 UP000001819 UP000001070 UP000008711 UP000008237 UP000007266 UP000215335 UP000000311 UP000007798 UP000009046 UP000019118 UP000030742 UP000053825 UP000242457 UP000005203 UP000076502 UP000005205 UP000078200 UP000027135 UP000092443 UP000078492 UP000078540 UP000078541 UP000092462 UP000078542 UP000053105 UP000235965 UP000079169 UP000015103 UP000075882
PRIDE
Interpro
SUPFAM
SSF64356
SSF64356
ProteinModelPortal
A0A2A4KA55
A0A194QF01
A0A2H1WNH3
I4DN83
A0A0N0PBX8
A0A1E1WN10
+ More
A0A437BBF2 A0A1B0CWA4 A0A1L8DDN2 B0WYR2 Q1HRE2 A0A182G698 A0A1Q3G471 U5ESJ9 T1DN85 A0A2M3ZB48 A0A2M4BWT2 W5JKT8 A0A182FJK1 A0A2M4AX15 A0A084VVH8 A0A182JH44 A0A182YH80 A0A182PSV7 A0A182LU20 A0A182SYP8 A0A1Y9HB63 A0A182KD93 A0A182U8F4 A0A182W5N2 A0A182XJX9 A0NCG5 A0A182RHL7 A0A182IAV9 W8CA05 A0A0L0C3W2 A0A034WCS9 A0A2Y9D318 A0A0K8VWZ5 A0A034WA63 A0A182N8G9 T1PBK4 A0A023ELS4 A0A1I8PT06 B3N0S0 B4M1S4 A0A026WTP1 B4R5T6 B4IF71 Q9VXG9 B4PXS7 B4L8R6 A0A1W4VHD7 A0A3B0KQ72 B4H0Z9 A0A0M5J5W1 Q29GI6 B4JKM4 B3NX45 E2BL30 D6WU90 A0A232FGQ6 E2AIS7 B4NEV3 E0W2J7 E9J0B0 V5G537 J3JWM4 A0A0L7RC96 A0A1B6K1J0 A0A2A3E5L9 A0A087ZY47 A0A1Y1MJQ4 A0A154P8Q8 A0A158NPX4 A0A1A9VRF7 A0A336MRA2 A0A067QTZ9 A0A1A9XDX3 A0A151IVK9 A0A195BET1 A0A195FH63 A0A1B0DNP4 A0A151IDD0 A0A1B6F8I2 A0A0N0BE69 A0A2J7R9A1 A0A1B6L944 A0A1S3DH01 A0A0A9XH50 A0A0P4VY36 R4WIN7 A0A224XVJ8 A0A0V0GC56 A0A069DR86 A0A1Q3G4B7 A0A0C9S187 T1HGV4 A0A1B6DH47 A0A182KJH1
A0A437BBF2 A0A1B0CWA4 A0A1L8DDN2 B0WYR2 Q1HRE2 A0A182G698 A0A1Q3G471 U5ESJ9 T1DN85 A0A2M3ZB48 A0A2M4BWT2 W5JKT8 A0A182FJK1 A0A2M4AX15 A0A084VVH8 A0A182JH44 A0A182YH80 A0A182PSV7 A0A182LU20 A0A182SYP8 A0A1Y9HB63 A0A182KD93 A0A182U8F4 A0A182W5N2 A0A182XJX9 A0NCG5 A0A182RHL7 A0A182IAV9 W8CA05 A0A0L0C3W2 A0A034WCS9 A0A2Y9D318 A0A0K8VWZ5 A0A034WA63 A0A182N8G9 T1PBK4 A0A023ELS4 A0A1I8PT06 B3N0S0 B4M1S4 A0A026WTP1 B4R5T6 B4IF71 Q9VXG9 B4PXS7 B4L8R6 A0A1W4VHD7 A0A3B0KQ72 B4H0Z9 A0A0M5J5W1 Q29GI6 B4JKM4 B3NX45 E2BL30 D6WU90 A0A232FGQ6 E2AIS7 B4NEV3 E0W2J7 E9J0B0 V5G537 J3JWM4 A0A0L7RC96 A0A1B6K1J0 A0A2A3E5L9 A0A087ZY47 A0A1Y1MJQ4 A0A154P8Q8 A0A158NPX4 A0A1A9VRF7 A0A336MRA2 A0A067QTZ9 A0A1A9XDX3 A0A151IVK9 A0A195BET1 A0A195FH63 A0A1B0DNP4 A0A151IDD0 A0A1B6F8I2 A0A0N0BE69 A0A2J7R9A1 A0A1B6L944 A0A1S3DH01 A0A0A9XH50 A0A0P4VY36 R4WIN7 A0A224XVJ8 A0A0V0GC56 A0A069DR86 A0A1Q3G4B7 A0A0C9S187 T1HGV4 A0A1B6DH47 A0A182KJH1
PDB
5EGI
E-value=6.21114e-72,
Score=686
Ontologies
GO
PANTHER
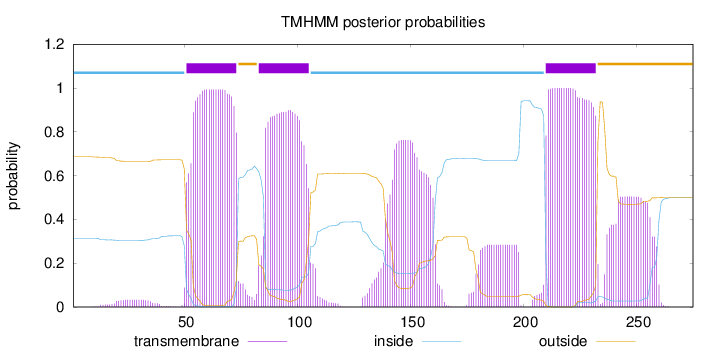
Topology
Length:
275
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
95.63419
Exp number, first 60 AAs:
9.33302
Total prob of N-in:
0.31348
inside
1 - 50
TMhelix
51 - 73
outside
74 - 82
TMhelix
83 - 105
inside
106 - 209
TMhelix
210 - 232
outside
233 - 275
Population Genetic Test Statistics
Pi
220.899958
Theta
186.382741
Tajima's D
0.4544
CLR
0.08698
CSRT
0.503424828758562
Interpretation
Uncertain
