Pre Gene Modal
BGIBMGA001960
Annotation
PREDICTED:_ufm1-specific_protease_1_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.891
Sequence
CDS
ATGTCAAATTTGTTGGTAAACATCCATGAATCTTTAGTATCAAAACAAACAGGAAAATCATATTTAATTAAAGGAAATTATGAATATTATCATTACTTGTGTGATGGATTTGATGATAGGGGCTGGGGTTGCGGTTATAGAACTCTTCAGACGATATGTTCCTGGATGAAATTAAATCATATGGATGTTGCTGTACCTTCAATACGAGAGATTCAATCGATACTGGTTGATTTGGAAGATAAACCGAAAACGTTTATTGGTTCGAGACAGTGGATTGGCAGTTTTGAGGTTTGCTTGGTAATTGATAAACTATTTGATGTTCCATGTAAAATTATTCATATAAACAAAGGAGACGATCTTAAAACTATTGTGGACGCCTTAGTCAAACACTTCATTGAGTACAGCAGTCCCGTAATGATGGGAGGAGATGTTGATTGCTCATCTAAAGGGATCATGGGCATTCATATTGGAGACCATGGTGCCAGTTTATTGGTGGTCGATCCACACTATGTTGGAAAACAACCACGTAAAAATTATCTACAAGACCATGGCTGGGTGAAGTGGCAACCACTCCATGATTTCCTGAGTTCGTCTTTCTATAATTTGTGTCTGCCTCAAGTTAAAGCAAAGAATGCGTGA
Protein
MSNLLVNIHESLVSKQTGKSYLIKGNYEYYHYLCDGFDDRGWGCGYRTLQTICSWMKLNHMDVAVPSIREIQSILVDLEDKPKTFIGSRQWIGSFEVCLVIDKLFDVPCKIIHINKGDDLKTIVDALVKHFIEYSSPVMMGGDVDCSSKGIMGIHIGDHGASLLVVDPHYVGKQPRKNYLQDHGWVKWQPLHDFLSSSFYNLCLPQVKAKNA
Summary
Uniprot
H9IXH7
A0A2W1BFZ4
A0A2A4JVD0
A0A2H1WLW2
A0A194QKM5
A0A437BBF1
+ More
A0A194RA51 A0A212EHS1 E2BZ42 A0A195E493 A0A151WUU5 A0A195CU28 A0A151I1Y1 F4WAD1 A0A158NKX2 E9IZ08 A0A2A3ESM1 A0A088AP77 A0A154PNM6 A0A026WQ29 A0A310SJC4 A0A0M8ZY74 A0A0L7QN85 A0A195FS90 A0A067RF08 A0A2J7QPL9 A0A1S3HFU5 A0A2J7QPL3 A0A2J7QPL6 A0A2J7QPL5 A0A0B6YP36 Q1HRD5 A0A182GSR3 A0A423T3Z7 A0A3Q2QLV9 A0A3P9PKL0 A0A3B3TZA6 A0A0P4W119 A0A3B3X1K8 A0A087YQ01 A0A182V2V0 A0A182LJJ5 Q7Q749 A0A3P9JFN8 A0A0K8TTA7 A0A3B3H6E1 A0A1A8JFA4 A0A3B5LVB8 H3AKJ3 A0A3P9MGG3 V4A9A1 A0A3Q2D6B8 M4AYP7 A0A084VZA5 A0A1L8DTS3 A0A182IPT8 A0A1L8DTJ4 A0A444SF88 A0A3B4YX24 A0A3Q1J4V1 F1QYX2 A0A3P8TU14 A0A3Q1C829 W4ZF44 A0A1A8BSP7 A0A1A8N803 A0A3Q1GB93 A0A3S1AH65 A0A1A8L771 A0A131XDB1 A0A3B3BB33 A0A0L8I4T6 A0A1A8A418 A0A3Q3QMT2 A0A0F8BDU0 A0A3Q3DBR9 A0A3Q3FNM3 A0A3N0YH91 L7LZC6 A0A147AYX3 A0A1Q3G4N7 A0A2U9BA67 A3KP75 A0A2I4CV46 A0A1A8ENV7 A0A3B4YC26 A0A2P2IC22 A0A3Q3RV21 A0A293MBH4 B0VYZ0 E9HCU4 A0A131YT39 A0A0K8RFM4 A0A3Q4GMM8 A0A1I8PTS7 A0A224YVG8 I3KYA8 G3Q8W3 A0A3P8QEJ6 A0A3P9B9T0
A0A194RA51 A0A212EHS1 E2BZ42 A0A195E493 A0A151WUU5 A0A195CU28 A0A151I1Y1 F4WAD1 A0A158NKX2 E9IZ08 A0A2A3ESM1 A0A088AP77 A0A154PNM6 A0A026WQ29 A0A310SJC4 A0A0M8ZY74 A0A0L7QN85 A0A195FS90 A0A067RF08 A0A2J7QPL9 A0A1S3HFU5 A0A2J7QPL3 A0A2J7QPL6 A0A2J7QPL5 A0A0B6YP36 Q1HRD5 A0A182GSR3 A0A423T3Z7 A0A3Q2QLV9 A0A3P9PKL0 A0A3B3TZA6 A0A0P4W119 A0A3B3X1K8 A0A087YQ01 A0A182V2V0 A0A182LJJ5 Q7Q749 A0A3P9JFN8 A0A0K8TTA7 A0A3B3H6E1 A0A1A8JFA4 A0A3B5LVB8 H3AKJ3 A0A3P9MGG3 V4A9A1 A0A3Q2D6B8 M4AYP7 A0A084VZA5 A0A1L8DTS3 A0A182IPT8 A0A1L8DTJ4 A0A444SF88 A0A3B4YX24 A0A3Q1J4V1 F1QYX2 A0A3P8TU14 A0A3Q1C829 W4ZF44 A0A1A8BSP7 A0A1A8N803 A0A3Q1GB93 A0A3S1AH65 A0A1A8L771 A0A131XDB1 A0A3B3BB33 A0A0L8I4T6 A0A1A8A418 A0A3Q3QMT2 A0A0F8BDU0 A0A3Q3DBR9 A0A3Q3FNM3 A0A3N0YH91 L7LZC6 A0A147AYX3 A0A1Q3G4N7 A0A2U9BA67 A3KP75 A0A2I4CV46 A0A1A8ENV7 A0A3B4YC26 A0A2P2IC22 A0A3Q3RV21 A0A293MBH4 B0VYZ0 E9HCU4 A0A131YT39 A0A0K8RFM4 A0A3Q4GMM8 A0A1I8PTS7 A0A224YVG8 I3KYA8 G3Q8W3 A0A3P8QEJ6 A0A3P9B9T0
Pubmed
19121390
28756777
26354079
22118469
20798317
21719571
+ More
21347285 21282665 24508170 30249741 24845553 17204158 17510324 26483478 20966253 12364791 14747013 17210077 17554307 26369729 9215903 23254933 23542700 24438588 23594743 28049606 29451363 25835551 25576852 21292972 26830274 25186727 28797301
21347285 21282665 24508170 30249741 24845553 17204158 17510324 26483478 20966253 12364791 14747013 17210077 17554307 26369729 9215903 23254933 23542700 24438588 23594743 28049606 29451363 25835551 25576852 21292972 26830274 25186727 28797301
EMBL
BABH01018149
KZ150261
PZC71736.1
NWSH01000608
PCG75352.1
ODYU01009554
+ More
SOQ54065.1 KQ459053 KPJ04006.1 RSAL01000096 RVE47736.1 KQ460436 KPJ14718.1 AGBW02014810 OWR41027.1 GL451555 EFN79044.1 KQ979701 KYN19674.1 KQ982718 KYQ51702.1 KQ977306 KYN03639.1 KQ976551 KYM80859.1 GL888048 EGI68814.1 ADTU01018996 GL767078 EFZ14192.1 KZ288193 PBC34041.1 KQ435007 KZC13453.1 KK107152 QOIP01000007 EZA57209.1 RLU20606.1 KQ759770 OAD62941.1 KQ435796 KOX73525.1 KQ414860 KOC60029.1 KQ981281 KYN43306.1 KK852678 KDR18663.1 NEVH01012088 PNF30527.1 PNF30524.1 PNF30529.1 PNF30526.1 HACG01011149 CEK58014.1 DQ440159 CH477822 ABF18192.1 EAT35923.1 JXUM01085296 JXUM01085297 KQ563494 KXJ73749.1 QCYY01002345 ROT71095.1 GDRN01097325 GDRN01097324 GDRN01097323 JAI59135.1 AYCK01010202 AAAB01008960 EAA11158.4 GDAI01000453 JAI17150.1 HAED01021880 HAEE01007372 SBR08633.1 AFYH01105154 KB201931 ESO93327.1 ATLV01018626 KE525239 KFB43299.1 GFDF01004304 JAV09780.1 GFDF01004311 JAV09773.1 SAUD01039222 RXG52079.1 BX005336 AAGJ04008818 AAGJ04008819 AAGJ04008820 HADZ01006278 SBP70219.1 HAEH01000596 SBR65200.1 RQTK01000003 RUS92030.1 HAEF01002946 SBR40328.1 GEFH01004935 JAP63646.1 KQ416547 KOF96516.1 HADY01010425 HAEJ01011857 SBP48910.1 KQ040943 KKF32754.1 RJVU01042551 ROL45615.1 GACK01008835 JAA56199.1 GCES01002632 JAR83691.1 GFDL01000319 JAV34726.1 CP026246 AWP00851.1 BC134191 AAI34192.1 HAEB01001624 SBQ48120.1 IACF01005976 LAB71559.1 GFWV01012619 MAA37348.1 DS231813 EDS25734.1 GL732621 EFX70413.1 GEDV01006907 JAP81650.1 GADI01003846 JAA69962.1 GFPF01010450 MAA21596.1 AERX01046678
SOQ54065.1 KQ459053 KPJ04006.1 RSAL01000096 RVE47736.1 KQ460436 KPJ14718.1 AGBW02014810 OWR41027.1 GL451555 EFN79044.1 KQ979701 KYN19674.1 KQ982718 KYQ51702.1 KQ977306 KYN03639.1 KQ976551 KYM80859.1 GL888048 EGI68814.1 ADTU01018996 GL767078 EFZ14192.1 KZ288193 PBC34041.1 KQ435007 KZC13453.1 KK107152 QOIP01000007 EZA57209.1 RLU20606.1 KQ759770 OAD62941.1 KQ435796 KOX73525.1 KQ414860 KOC60029.1 KQ981281 KYN43306.1 KK852678 KDR18663.1 NEVH01012088 PNF30527.1 PNF30524.1 PNF30529.1 PNF30526.1 HACG01011149 CEK58014.1 DQ440159 CH477822 ABF18192.1 EAT35923.1 JXUM01085296 JXUM01085297 KQ563494 KXJ73749.1 QCYY01002345 ROT71095.1 GDRN01097325 GDRN01097324 GDRN01097323 JAI59135.1 AYCK01010202 AAAB01008960 EAA11158.4 GDAI01000453 JAI17150.1 HAED01021880 HAEE01007372 SBR08633.1 AFYH01105154 KB201931 ESO93327.1 ATLV01018626 KE525239 KFB43299.1 GFDF01004304 JAV09780.1 GFDF01004311 JAV09773.1 SAUD01039222 RXG52079.1 BX005336 AAGJ04008818 AAGJ04008819 AAGJ04008820 HADZ01006278 SBP70219.1 HAEH01000596 SBR65200.1 RQTK01000003 RUS92030.1 HAEF01002946 SBR40328.1 GEFH01004935 JAP63646.1 KQ416547 KOF96516.1 HADY01010425 HAEJ01011857 SBP48910.1 KQ040943 KKF32754.1 RJVU01042551 ROL45615.1 GACK01008835 JAA56199.1 GCES01002632 JAR83691.1 GFDL01000319 JAV34726.1 CP026246 AWP00851.1 BC134191 AAI34192.1 HAEB01001624 SBQ48120.1 IACF01005976 LAB71559.1 GFWV01012619 MAA37348.1 DS231813 EDS25734.1 GL732621 EFX70413.1 GEDV01006907 JAP81650.1 GADI01003846 JAA69962.1 GFPF01010450 MAA21596.1 AERX01046678
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000007151
+ More
UP000008237 UP000078492 UP000075809 UP000078542 UP000078540 UP000007755 UP000005205 UP000242457 UP000005203 UP000076502 UP000053097 UP000279307 UP000053105 UP000053825 UP000078541 UP000027135 UP000235965 UP000085678 UP000008820 UP000069940 UP000249989 UP000283509 UP000265000 UP000242638 UP000261500 UP000261480 UP000028760 UP000075903 UP000075882 UP000007062 UP000265200 UP000001038 UP000261380 UP000008672 UP000265180 UP000030746 UP000265020 UP000002852 UP000030765 UP000075880 UP000288706 UP000261400 UP000265040 UP000000437 UP000265080 UP000257160 UP000007110 UP000257200 UP000271974 UP000261560 UP000053454 UP000261600 UP000264820 UP000264800 UP000246464 UP000192220 UP000261360 UP000261640 UP000002320 UP000000305 UP000261580 UP000095300 UP000005207 UP000007635 UP000265100 UP000265160
UP000008237 UP000078492 UP000075809 UP000078542 UP000078540 UP000007755 UP000005205 UP000242457 UP000005203 UP000076502 UP000053097 UP000279307 UP000053105 UP000053825 UP000078541 UP000027135 UP000235965 UP000085678 UP000008820 UP000069940 UP000249989 UP000283509 UP000265000 UP000242638 UP000261500 UP000261480 UP000028760 UP000075903 UP000075882 UP000007062 UP000265200 UP000001038 UP000261380 UP000008672 UP000265180 UP000030746 UP000265020 UP000002852 UP000030765 UP000075880 UP000288706 UP000261400 UP000265040 UP000000437 UP000265080 UP000257160 UP000007110 UP000257200 UP000271974 UP000261560 UP000053454 UP000261600 UP000264820 UP000264800 UP000246464 UP000192220 UP000261360 UP000261640 UP000002320 UP000000305 UP000261580 UP000095300 UP000005207 UP000007635 UP000265100 UP000265160
PRIDE
Pfam
PF07910 Peptidase_C78
SUPFAM
SSF54001
SSF54001
ProteinModelPortal
H9IXH7
A0A2W1BFZ4
A0A2A4JVD0
A0A2H1WLW2
A0A194QKM5
A0A437BBF1
+ More
A0A194RA51 A0A212EHS1 E2BZ42 A0A195E493 A0A151WUU5 A0A195CU28 A0A151I1Y1 F4WAD1 A0A158NKX2 E9IZ08 A0A2A3ESM1 A0A088AP77 A0A154PNM6 A0A026WQ29 A0A310SJC4 A0A0M8ZY74 A0A0L7QN85 A0A195FS90 A0A067RF08 A0A2J7QPL9 A0A1S3HFU5 A0A2J7QPL3 A0A2J7QPL6 A0A2J7QPL5 A0A0B6YP36 Q1HRD5 A0A182GSR3 A0A423T3Z7 A0A3Q2QLV9 A0A3P9PKL0 A0A3B3TZA6 A0A0P4W119 A0A3B3X1K8 A0A087YQ01 A0A182V2V0 A0A182LJJ5 Q7Q749 A0A3P9JFN8 A0A0K8TTA7 A0A3B3H6E1 A0A1A8JFA4 A0A3B5LVB8 H3AKJ3 A0A3P9MGG3 V4A9A1 A0A3Q2D6B8 M4AYP7 A0A084VZA5 A0A1L8DTS3 A0A182IPT8 A0A1L8DTJ4 A0A444SF88 A0A3B4YX24 A0A3Q1J4V1 F1QYX2 A0A3P8TU14 A0A3Q1C829 W4ZF44 A0A1A8BSP7 A0A1A8N803 A0A3Q1GB93 A0A3S1AH65 A0A1A8L771 A0A131XDB1 A0A3B3BB33 A0A0L8I4T6 A0A1A8A418 A0A3Q3QMT2 A0A0F8BDU0 A0A3Q3DBR9 A0A3Q3FNM3 A0A3N0YH91 L7LZC6 A0A147AYX3 A0A1Q3G4N7 A0A2U9BA67 A3KP75 A0A2I4CV46 A0A1A8ENV7 A0A3B4YC26 A0A2P2IC22 A0A3Q3RV21 A0A293MBH4 B0VYZ0 E9HCU4 A0A131YT39 A0A0K8RFM4 A0A3Q4GMM8 A0A1I8PTS7 A0A224YVG8 I3KYA8 G3Q8W3 A0A3P8QEJ6 A0A3P9B9T0
A0A194RA51 A0A212EHS1 E2BZ42 A0A195E493 A0A151WUU5 A0A195CU28 A0A151I1Y1 F4WAD1 A0A158NKX2 E9IZ08 A0A2A3ESM1 A0A088AP77 A0A154PNM6 A0A026WQ29 A0A310SJC4 A0A0M8ZY74 A0A0L7QN85 A0A195FS90 A0A067RF08 A0A2J7QPL9 A0A1S3HFU5 A0A2J7QPL3 A0A2J7QPL6 A0A2J7QPL5 A0A0B6YP36 Q1HRD5 A0A182GSR3 A0A423T3Z7 A0A3Q2QLV9 A0A3P9PKL0 A0A3B3TZA6 A0A0P4W119 A0A3B3X1K8 A0A087YQ01 A0A182V2V0 A0A182LJJ5 Q7Q749 A0A3P9JFN8 A0A0K8TTA7 A0A3B3H6E1 A0A1A8JFA4 A0A3B5LVB8 H3AKJ3 A0A3P9MGG3 V4A9A1 A0A3Q2D6B8 M4AYP7 A0A084VZA5 A0A1L8DTS3 A0A182IPT8 A0A1L8DTJ4 A0A444SF88 A0A3B4YX24 A0A3Q1J4V1 F1QYX2 A0A3P8TU14 A0A3Q1C829 W4ZF44 A0A1A8BSP7 A0A1A8N803 A0A3Q1GB93 A0A3S1AH65 A0A1A8L771 A0A131XDB1 A0A3B3BB33 A0A0L8I4T6 A0A1A8A418 A0A3Q3QMT2 A0A0F8BDU0 A0A3Q3DBR9 A0A3Q3FNM3 A0A3N0YH91 L7LZC6 A0A147AYX3 A0A1Q3G4N7 A0A2U9BA67 A3KP75 A0A2I4CV46 A0A1A8ENV7 A0A3B4YC26 A0A2P2IC22 A0A3Q3RV21 A0A293MBH4 B0VYZ0 E9HCU4 A0A131YT39 A0A0K8RFM4 A0A3Q4GMM8 A0A1I8PTS7 A0A224YVG8 I3KYA8 G3Q8W3 A0A3P8QEJ6 A0A3P9B9T0
PDB
5EJJ
E-value=8.85634e-43,
Score=433
Ontologies
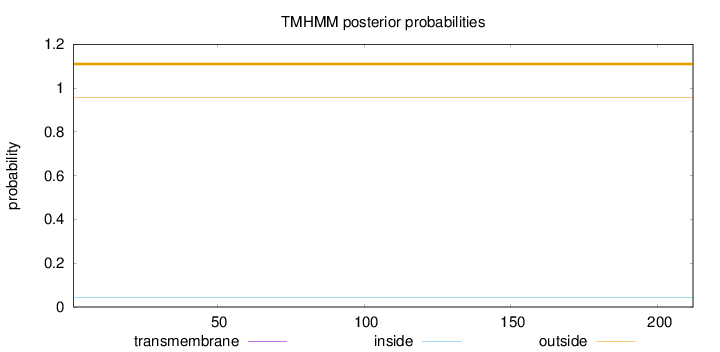
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0029
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04192
outside
1 - 212
Population Genetic Test Statistics
Pi
175.285163
Theta
160.529573
Tajima's D
0.64532
CLR
0
CSRT
0.559422028898555
Interpretation
Uncertain
