Pre Gene Modal
BGIBMGA001859
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_dynein_heavy_chain_10?_axonemal-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.526 PlasmaMembrane Reliability : 1.11
Sequence
CDS
ATGCTTGTACCTAGTAATCACTCGTGTCTGATAGAATCTGTTTTCTGTGGATTCGAACAGGTGATGGAAAAACTGGAACTGGCCGAAGCAACCACAAAAGATATAGAAAAACTAAGGGACGGCTACCGTCCGGTGGCGAAGCGCGGCTCCATACTGTTCTTCGTTCTATCCGACATGGCGGGCGTGAACAGCATGTACCAATACTCGCTGTCTTCATATCTAGACGTTTTCTCGTTTTCGTTGCGGAAAGCGATGCCGAACGTGATTTTAGTGAAGAGACTTAAGAACATAATTGATATGCTCACTAAAAACGTCTACGACTACGGCTGTACAGGTATATTCTTGCAACGGGCTATTACACATAAACTGTAG
Protein
MLVPSNHSCLIESVFCGFEQVMEKLELAEATTKDIEKLRDGYRPVAKRGSILFFVLSDMAGVNSMYQYSLSSYLDVFSFSLRKAMPNVILVKRLKNIIDMLTKNVYDYGCTGIFLQRAITHKL
Summary
Uniprot
A0A3S2LP16
H9IX76
A0A2W1BD47
A0A194QEV0
A0A194RB51
A0A3S2LZK1
+ More
A0A212EHR2 A0A2H1VR18 U4U1Q7 N6TR49 A0A183XAH2 A0A3P8KBE1 W5MFQ0 D6WU89 A0A0B7A7I1 K7FM63 A0A452I0T0 A0A452I0W3 A0A1S3PFH5 A0A1S3PEP0 A0A310SST6 A0A151NS45 A0A3Q0RTW8 C3XV30 A0A1U8CX33 M7BIQ2 A0A034V0L7 A0A183Q1F5 A0A154PNN1 A0A2A3ERY9 A0A1S3INJ4 A0A060W723 A0A3P9A2D6 A0A401Q7P6 A0A2H1CC55 A0A088ANX7 A0A0L0CEV9 A0A402F8C2 A0A1A9XJK5 A0A1S3HMR6 A0A2R2MKY4 F7DW33 A0A2G8KKM7 W5K6M6 A0A3S1HDS1 A0A1B0C738 A0A1L8I0W3 H9GM01 A0A3P8RA28 A0A3B3XVX4 A0A1A9X2L4 A0A2G9SBI3 A0A2D0PL14 A0A2D0PNR9 A0A1D2MX88 A0A3Q4MJC5 A0A3B5MHF8 A0A1I8NF00 H0WZ43 A0A3P9CLL7 A0A3B4GFI9 A0A3Q2WFN5 A0A3B5QTB2 A0A0L8GLR3 A0A2T7PHE2 A0A0L8GLN5 A0A3B3QK43 A0A0J7L4X0 A0A1S3W223 A0A0N0U4V3 A0A315UWI8 A0A087XTH1 A0A1W4ZUV9 A0A0N8JY00 A0A1S3FRL5 E2BZ36 A0A3B4YPF3 A0A3B3TNK4 A0A3B4E236 A0A1B0G3I6 R7UWA6 F7CP57 A0A210Q0K8 A0A430Q0A1 A0A1A9ZFQ2 A0A151I1T7 A0A1A9UNN9 A0A3N0XJC6 A0A182IR22 L5KS76 A0A2C9JWZ8 A0A2C9JWZ0 H3BAL0 A0A095AHA3 A0A3B4UD57 A0A2T7PHE3 T1JML4 A0A3Q3E3F5 A0A0M4EPR1 W4ZEE6
A0A212EHR2 A0A2H1VR18 U4U1Q7 N6TR49 A0A183XAH2 A0A3P8KBE1 W5MFQ0 D6WU89 A0A0B7A7I1 K7FM63 A0A452I0T0 A0A452I0W3 A0A1S3PFH5 A0A1S3PEP0 A0A310SST6 A0A151NS45 A0A3Q0RTW8 C3XV30 A0A1U8CX33 M7BIQ2 A0A034V0L7 A0A183Q1F5 A0A154PNN1 A0A2A3ERY9 A0A1S3INJ4 A0A060W723 A0A3P9A2D6 A0A401Q7P6 A0A2H1CC55 A0A088ANX7 A0A0L0CEV9 A0A402F8C2 A0A1A9XJK5 A0A1S3HMR6 A0A2R2MKY4 F7DW33 A0A2G8KKM7 W5K6M6 A0A3S1HDS1 A0A1B0C738 A0A1L8I0W3 H9GM01 A0A3P8RA28 A0A3B3XVX4 A0A1A9X2L4 A0A2G9SBI3 A0A2D0PL14 A0A2D0PNR9 A0A1D2MX88 A0A3Q4MJC5 A0A3B5MHF8 A0A1I8NF00 H0WZ43 A0A3P9CLL7 A0A3B4GFI9 A0A3Q2WFN5 A0A3B5QTB2 A0A0L8GLR3 A0A2T7PHE2 A0A0L8GLN5 A0A3B3QK43 A0A0J7L4X0 A0A1S3W223 A0A0N0U4V3 A0A315UWI8 A0A087XTH1 A0A1W4ZUV9 A0A0N8JY00 A0A1S3FRL5 E2BZ36 A0A3B4YPF3 A0A3B3TNK4 A0A3B4E236 A0A1B0G3I6 R7UWA6 F7CP57 A0A210Q0K8 A0A430Q0A1 A0A1A9ZFQ2 A0A151I1T7 A0A1A9UNN9 A0A3N0XJC6 A0A182IR22 L5KS76 A0A2C9JWZ8 A0A2C9JWZ0 H3BAL0 A0A095AHA3 A0A3B4UD57 A0A2T7PHE3 T1JML4 A0A3Q3E3F5 A0A0M4EPR1 W4ZEE6
Pubmed
19121390
28756777
26354079
22118469
23537049
18362917
+ More
19820115 17381049 28562605 22293439 18563158 23624526 25348373 24755649 25069045 30297745 26108605 26383154 20431018 29023486 25329095 27762356 21881562 27289101 25186727 25315136 23542700 29240929 29703783 20798317 23254933 18464734 28812685 23258410 15562597 9215903 22246508
19820115 17381049 28562605 22293439 18563158 23624526 25348373 24755649 25069045 30297745 26108605 26383154 20431018 29023486 25329095 27762356 21881562 27289101 25186727 25315136 23542700 29240929 29703783 20798317 23254933 18464734 28812685 23258410 15562597 9215903 22246508
EMBL
RSAL01001995
RVE40608.1
BABH01018149
BABH01018150
BABH01018151
BABH01018152
+ More
BABH01018153 BABH01018154 BABH01018155 BABH01018156 BABH01018157 BABH01018158 KZ150261 PZC71734.1 KQ459053 KPJ04007.1 KQ460436 KPJ14719.1 RSAL01000096 RVE47735.1 AGBW02014810 OWR41026.1 ODYU01003927 SOQ43291.1 KB631802 ERL86248.1 APGK01057806 KB741282 ENN70796.1 UZAO01134930 VDQ17255.1 AHAT01005142 AHAT01005143 KQ971352 EFA06783.2 HACG01030059 CEK76924.1 AGCU01061902 AGCU01061903 AGCU01061904 AGCU01061905 AGCU01061906 AGCU01061907 AGCU01061908 AGCU01061909 AGCU01061910 AGCU01061911 KQ759770 OAD62940.1 AKHW03002185 KYO39614.1 GG666468 EEN67956.1 KB523138 EMP37074.1 GAKP01023284 JAC35672.1 UZAL01044444 VDP82477.1 KQ435007 KZC13452.1 KZ288193 PBC34042.1 FR904417 CDQ62791.1 BFAA01020178 GCB81414.1 KZ429962 PIS85059.1 JRES01000501 KNC30752.1 BDOT01000138 GCF52688.1 AAMC01042923 AAMC01042924 MRZV01000518 PIK48527.1 RQTK01000590 RUS77325.1 JXJN01027505 JXJN01027506 JXJN01027507 CM004466 OCU02010.1 KV926115 PIO37502.1 LJIJ01000456 ODM97295.1 AAQR03124676 AAQR03124677 AAQR03124678 KQ421273 KOF77888.1 PZQS01000004 PVD32838.1 KOF77887.1 LBMM01000749 KMQ97578.1 KQ435796 KOX73524.1 NHOQ01002573 PWA15723.1 AYCK01007726 JARO02006505 KPP65153.1 GL451555 EFN79038.1 CCAG010016326 CCAG010016327 AMQN01006005 KB297391 ELU10552.1 NEDP02005302 OWF42267.1 QMKO01003563 RTG81154.1 KQ976551 KYM80860.1 RJVU01073043 ROI27764.1 KB030576 ELK14267.1 AFYH01016331 AFYH01016332 AFYH01016333 AFYH01016334 AFYH01016335 AFYH01016336 AFYH01016337 AFYH01016338 AFYH01016339 AFYH01016340 KL250542 KGB33311.1 PVD32839.1 JH431878 CP012526 ALC46147.1 AAGJ04084865 AAGJ04084866 AAGJ04084867 AAGJ04084868
BABH01018153 BABH01018154 BABH01018155 BABH01018156 BABH01018157 BABH01018158 KZ150261 PZC71734.1 KQ459053 KPJ04007.1 KQ460436 KPJ14719.1 RSAL01000096 RVE47735.1 AGBW02014810 OWR41026.1 ODYU01003927 SOQ43291.1 KB631802 ERL86248.1 APGK01057806 KB741282 ENN70796.1 UZAO01134930 VDQ17255.1 AHAT01005142 AHAT01005143 KQ971352 EFA06783.2 HACG01030059 CEK76924.1 AGCU01061902 AGCU01061903 AGCU01061904 AGCU01061905 AGCU01061906 AGCU01061907 AGCU01061908 AGCU01061909 AGCU01061910 AGCU01061911 KQ759770 OAD62940.1 AKHW03002185 KYO39614.1 GG666468 EEN67956.1 KB523138 EMP37074.1 GAKP01023284 JAC35672.1 UZAL01044444 VDP82477.1 KQ435007 KZC13452.1 KZ288193 PBC34042.1 FR904417 CDQ62791.1 BFAA01020178 GCB81414.1 KZ429962 PIS85059.1 JRES01000501 KNC30752.1 BDOT01000138 GCF52688.1 AAMC01042923 AAMC01042924 MRZV01000518 PIK48527.1 RQTK01000590 RUS77325.1 JXJN01027505 JXJN01027506 JXJN01027507 CM004466 OCU02010.1 KV926115 PIO37502.1 LJIJ01000456 ODM97295.1 AAQR03124676 AAQR03124677 AAQR03124678 KQ421273 KOF77888.1 PZQS01000004 PVD32838.1 KOF77887.1 LBMM01000749 KMQ97578.1 KQ435796 KOX73524.1 NHOQ01002573 PWA15723.1 AYCK01007726 JARO02006505 KPP65153.1 GL451555 EFN79038.1 CCAG010016326 CCAG010016327 AMQN01006005 KB297391 ELU10552.1 NEDP02005302 OWF42267.1 QMKO01003563 RTG81154.1 KQ976551 KYM80860.1 RJVU01073043 ROI27764.1 KB030576 ELK14267.1 AFYH01016331 AFYH01016332 AFYH01016333 AFYH01016334 AFYH01016335 AFYH01016336 AFYH01016337 AFYH01016338 AFYH01016339 AFYH01016340 KL250542 KGB33311.1 PVD32839.1 JH431878 CP012526 ALC46147.1 AAGJ04084865 AAGJ04084866 AAGJ04084867 AAGJ04084868
Proteomes
UP000283053
UP000005204
UP000053268
UP000053240
UP000007151
UP000030742
+ More
UP000019118 UP000050795 UP000018468 UP000007266 UP000007267 UP000291020 UP000087266 UP000050525 UP000261340 UP000001554 UP000189705 UP000031443 UP000050791 UP000076502 UP000242457 UP000085678 UP000193380 UP000265140 UP000288216 UP000005203 UP000037069 UP000288954 UP000092443 UP000008143 UP000230750 UP000018467 UP000271974 UP000092460 UP000186698 UP000001646 UP000265100 UP000261480 UP000091820 UP000221080 UP000094527 UP000261580 UP000261380 UP000095301 UP000005225 UP000265160 UP000261460 UP000264840 UP000002852 UP000053454 UP000245119 UP000261540 UP000036403 UP000079721 UP000053105 UP000028760 UP000192224 UP000034805 UP000081671 UP000008237 UP000261360 UP000261500 UP000261440 UP000092444 UP000014760 UP000002279 UP000242188 UP000092445 UP000078540 UP000078200 UP000075880 UP000010552 UP000076420 UP000008672 UP000261420 UP000261660 UP000092553 UP000007110
UP000019118 UP000050795 UP000018468 UP000007266 UP000007267 UP000291020 UP000087266 UP000050525 UP000261340 UP000001554 UP000189705 UP000031443 UP000050791 UP000076502 UP000242457 UP000085678 UP000193380 UP000265140 UP000288216 UP000005203 UP000037069 UP000288954 UP000092443 UP000008143 UP000230750 UP000018467 UP000271974 UP000092460 UP000186698 UP000001646 UP000265100 UP000261480 UP000091820 UP000221080 UP000094527 UP000261580 UP000261380 UP000095301 UP000005225 UP000265160 UP000261460 UP000264840 UP000002852 UP000053454 UP000245119 UP000261540 UP000036403 UP000079721 UP000053105 UP000028760 UP000192224 UP000034805 UP000081671 UP000008237 UP000261360 UP000261500 UP000261440 UP000092444 UP000014760 UP000002279 UP000242188 UP000092445 UP000078540 UP000078200 UP000075880 UP000010552 UP000076420 UP000008672 UP000261420 UP000261660 UP000092553 UP000007110
PRIDE
Pfam
Interpro
IPR042222
Dynein_2_N
+ More
IPR041589 DNAH3_AAA_lid_1
IPR035706 AAA_9
IPR024317 Dynein_heavy_chain_D4_dom
IPR024743 Dynein_HC_stalk
IPR027417 P-loop_NTPase
IPR041466 Dynein_AAA5_ext
IPR003593 AAA+_ATPase
IPR013594 Dynein_heavy_dom-1
IPR042219 AAA_lid_11_sf
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR035699 AAA_6
IPR041228 Dynein_C
IPR042228 Dynein_2_C
IPR041658 AAA_lid_11
IPR026983 DHC_fam
IPR036465 vWFA_dom_sf
IPR011677 DUF1619
IPR004600 TFIIH_Tfb4/GTF2H3
IPR002490 V-ATPase_116kDa_su
IPR011704 ATPase_dyneun-rel_AAA
IPR036305 RGS_sf
IPR016137 RGS
IPR037719 AKAP10_AKB_dom
IPR041589 DNAH3_AAA_lid_1
IPR035706 AAA_9
IPR024317 Dynein_heavy_chain_D4_dom
IPR024743 Dynein_HC_stalk
IPR027417 P-loop_NTPase
IPR041466 Dynein_AAA5_ext
IPR003593 AAA+_ATPase
IPR013594 Dynein_heavy_dom-1
IPR042219 AAA_lid_11_sf
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR035699 AAA_6
IPR041228 Dynein_C
IPR042228 Dynein_2_C
IPR041658 AAA_lid_11
IPR026983 DHC_fam
IPR036465 vWFA_dom_sf
IPR011677 DUF1619
IPR004600 TFIIH_Tfb4/GTF2H3
IPR002490 V-ATPase_116kDa_su
IPR011704 ATPase_dyneun-rel_AAA
IPR036305 RGS_sf
IPR016137 RGS
IPR037719 AKAP10_AKB_dom
Gene 3D
ProteinModelPortal
A0A3S2LP16
H9IX76
A0A2W1BD47
A0A194QEV0
A0A194RB51
A0A3S2LZK1
+ More
A0A212EHR2 A0A2H1VR18 U4U1Q7 N6TR49 A0A183XAH2 A0A3P8KBE1 W5MFQ0 D6WU89 A0A0B7A7I1 K7FM63 A0A452I0T0 A0A452I0W3 A0A1S3PFH5 A0A1S3PEP0 A0A310SST6 A0A151NS45 A0A3Q0RTW8 C3XV30 A0A1U8CX33 M7BIQ2 A0A034V0L7 A0A183Q1F5 A0A154PNN1 A0A2A3ERY9 A0A1S3INJ4 A0A060W723 A0A3P9A2D6 A0A401Q7P6 A0A2H1CC55 A0A088ANX7 A0A0L0CEV9 A0A402F8C2 A0A1A9XJK5 A0A1S3HMR6 A0A2R2MKY4 F7DW33 A0A2G8KKM7 W5K6M6 A0A3S1HDS1 A0A1B0C738 A0A1L8I0W3 H9GM01 A0A3P8RA28 A0A3B3XVX4 A0A1A9X2L4 A0A2G9SBI3 A0A2D0PL14 A0A2D0PNR9 A0A1D2MX88 A0A3Q4MJC5 A0A3B5MHF8 A0A1I8NF00 H0WZ43 A0A3P9CLL7 A0A3B4GFI9 A0A3Q2WFN5 A0A3B5QTB2 A0A0L8GLR3 A0A2T7PHE2 A0A0L8GLN5 A0A3B3QK43 A0A0J7L4X0 A0A1S3W223 A0A0N0U4V3 A0A315UWI8 A0A087XTH1 A0A1W4ZUV9 A0A0N8JY00 A0A1S3FRL5 E2BZ36 A0A3B4YPF3 A0A3B3TNK4 A0A3B4E236 A0A1B0G3I6 R7UWA6 F7CP57 A0A210Q0K8 A0A430Q0A1 A0A1A9ZFQ2 A0A151I1T7 A0A1A9UNN9 A0A3N0XJC6 A0A182IR22 L5KS76 A0A2C9JWZ8 A0A2C9JWZ0 H3BAL0 A0A095AHA3 A0A3B4UD57 A0A2T7PHE3 T1JML4 A0A3Q3E3F5 A0A0M4EPR1 W4ZEE6
A0A212EHR2 A0A2H1VR18 U4U1Q7 N6TR49 A0A183XAH2 A0A3P8KBE1 W5MFQ0 D6WU89 A0A0B7A7I1 K7FM63 A0A452I0T0 A0A452I0W3 A0A1S3PFH5 A0A1S3PEP0 A0A310SST6 A0A151NS45 A0A3Q0RTW8 C3XV30 A0A1U8CX33 M7BIQ2 A0A034V0L7 A0A183Q1F5 A0A154PNN1 A0A2A3ERY9 A0A1S3INJ4 A0A060W723 A0A3P9A2D6 A0A401Q7P6 A0A2H1CC55 A0A088ANX7 A0A0L0CEV9 A0A402F8C2 A0A1A9XJK5 A0A1S3HMR6 A0A2R2MKY4 F7DW33 A0A2G8KKM7 W5K6M6 A0A3S1HDS1 A0A1B0C738 A0A1L8I0W3 H9GM01 A0A3P8RA28 A0A3B3XVX4 A0A1A9X2L4 A0A2G9SBI3 A0A2D0PL14 A0A2D0PNR9 A0A1D2MX88 A0A3Q4MJC5 A0A3B5MHF8 A0A1I8NF00 H0WZ43 A0A3P9CLL7 A0A3B4GFI9 A0A3Q2WFN5 A0A3B5QTB2 A0A0L8GLR3 A0A2T7PHE2 A0A0L8GLN5 A0A3B3QK43 A0A0J7L4X0 A0A1S3W223 A0A0N0U4V3 A0A315UWI8 A0A087XTH1 A0A1W4ZUV9 A0A0N8JY00 A0A1S3FRL5 E2BZ36 A0A3B4YPF3 A0A3B3TNK4 A0A3B4E236 A0A1B0G3I6 R7UWA6 F7CP57 A0A210Q0K8 A0A430Q0A1 A0A1A9ZFQ2 A0A151I1T7 A0A1A9UNN9 A0A3N0XJC6 A0A182IR22 L5KS76 A0A2C9JWZ8 A0A2C9JWZ0 H3BAL0 A0A095AHA3 A0A3B4UD57 A0A2T7PHE3 T1JML4 A0A3Q3E3F5 A0A0M4EPR1 W4ZEE6
PDB
4RH7
E-value=1.98077e-10,
Score=151
Ontologies
GO
PANTHER
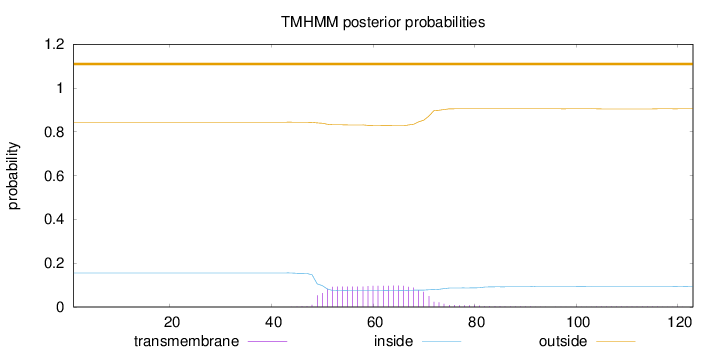
Topology
Length:
123
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.18952
Exp number, first 60 AAs:
1.06377
Total prob of N-in:
0.15579
outside
1 - 123
Population Genetic Test Statistics
Pi
213.362586
Theta
117.734547
Tajima's D
2.342867
CLR
0.000837
CSRT
0.927653617319134
Interpretation
Uncertain
