Pre Gene Modal
BGIBMGA001859
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_dynein_heavy_chain_10?_axonemal-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.477 Golgi Reliability : 1.586
Sequence
CDS
ATGATCGGTATGATACCGGCTCTGTTCGGTGATGATGAGAAGGACAGCATCATCAACTCTGTGAGAAACGACAGCAGCGATGCCGGATACGGGATAGCTAAGGATGCAGTTTGGAACTTCTTCTGCAACAGATGCGCGGACAATCTACACGTAGTGCTGTCGATGTCTCCGAGCGGAGACATTCTTAGAAACAGGTGTCGGAGTTTTCCTGGTCTAGTGAATAACACGACAATCGACTGGCAATTCCCTTGGCCGAAGCAGGCGTTGTTAGCTGTTGCTAACGTGTTTCTCGCTGATGTTCAGAAAATTCCAGAAGAATTTCGTCCCATAATAGTGGAGCATGTAGTCCACGTGCACATGTCGGTAGCACGATACTCTGCAGAGTTCCTGCTTCGACTTCGAAGAAACAACTACGTTACGCCCAAACATTACATGGATTTTCTAACCAATTATTTGGCTCTTCTAAATGAAAAAGACGCTTTCATAGTCGCTCAGTGTGAACGATTAAAAGGAGGGCTTGCTAAAATAGCCGAAGCAAATGTTCAGTTAGAAGACTTGAATGCGAAACTCGCAGTCCAGAAGGTTATCGTCGCCGAACAAACTAAGGAATGTGAAATATTGCTTAAAGAAATTTCTACAGCTACTGAAGCCGCAGTAGCAAAACAGCAAGTGGCGGGAGTGAAGTCGGTTGAGATCACCGAACAGAGCAAGGTCATCGCCGTGGAGAAGGCTGATGCTGAGGAGGCGTTGTCTGCTGCGTTGCCTGCACTAGAAGCTGCAAGATTGGCCCTTGCTGATTTGGACAAGAACGATATTACTGAAATTCGATCTTTTGCAACACCCCCTGAAGCTGTACAGGTGGTGTGTGAATGCGTCGTAATAATCAGAGGAATTAAAGATGTAAGTTGGAAAGGCGCCAAGGGAATGATGGCAGATCCAAATTTCTTGCGGAATCTTCAAGAGATGAACTGCGATCTTATCACTCAGGCACAAGTTAAGGCGGTAAAAACGCACATGAAAAAGAGTAAAAAACTGGATACGATGCAGCAGATTAGCAAAGCTGGATATGGACTACTAAAGTTTGTCACTGCAGTATTGGGCTATTGTGCCGTCTACAAAGAAGTCAAACCAAAAAAAGAGAAAGTAGAAGCACTAGAGAAAGAATATTCGGAAGCTGTCAACTATTTGGCTTCCCTCAATAGAGAAATCGACCGACTACAGAAGACTTTAGACGGACTAAACAATCGTTACGAAACGGCCATGATGAGAAGACAGGAACTTCAAGAAGAAACCGATCTGATGATGCGGAGATTGGTGGCAGCAGATAAACTAATGTCTGGACTGTCTAGTGAGCAGAAAAGGTGGACAGAAGATTTGGCAGCCTTATATGTAGAACAATCGAGGCTTATAGGAAACTGTTTGTTGGCAACGTCTTTCTTGAGCTACACCGGTCCTTTCTCGTTTTCATTTCGACAAACTATGATTTACGAGGATTGGTTGGGCGATGTTATGGAGAGGGGAATACCGTTGACTCTACCCTTTACAATTGAACGAAATTTGACTAATGAAGTTGAAGTCAGCGGGTGGAATAGTGAAGGTCTACCTCCAGATGAGTTGTCAGTCCAAAACGGAATACTAACAACAAGAGCCTCGAGGTTTCCATTGTGCATCGACCCTCAAACGCAAGCTCTCACATGGATAAAAAAGAAGGAGGCTAAAAATAATCTAAAGGTCCTATCGTTTAACGATCCACAATTCTTGAGGCAATTGGAGATGGCGATCAAGTACGGAATGCCGGTTCTGTTTCAAGACGTAAATGAGTATATCGATCCAGTTGTAGATAATGTTTTGGAAAAGAATATCAAAGTGGAAAGCGGTCGGACATTCGTGATGCTAGGTAGCACGGAAGTAGACTACGACCCTAACTTCCGCATGTACCTGACCACGAAGCTCGCCAACCCTCAATTCAACCCGGCTGCGTATGCGAAAGCCGTTGTTATTAACTACACAGTCACCGTTCAGGGTCTCGAAGATCAGCTTCTCAGCGTAGTAGTACGCGCAGAACGTTCAGATCTTGAGGAGCAGCGGGAAAGCCTCATCATCGAGACCAGCGCGAACAAGAGTTTACTTTCTGGACTGGAGGACTCTCTGCTCAGGGAGCTGGCCACGTCGACCGGTAACATGCTGGATAATGTTGAACTGGTCAATACTTTGGAAAACACCAAGTCTAAAGCTGCTGAGGTGAGGAAATACTGA
Protein
MIGMIPALFGDDEKDSIINSVRNDSSDAGYGIAKDAVWNFFCNRCADNLHVVLSMSPSGDILRNRCRSFPGLVNNTTIDWQFPWPKQALLAVANVFLADVQKIPEEFRPIIVEHVVHVHMSVARYSAEFLLRLRRNNYVTPKHYMDFLTNYLALLNEKDAFIVAQCERLKGGLAKIAEANVQLEDLNAKLAVQKVIVAEQTKECEILLKEISTATEAAVAKQQVAGVKSVEITEQSKVIAVEKADAEEALSAALPALEAARLALADLDKNDITEIRSFATPPEAVQVVCECVVIIRGIKDVSWKGAKGMMADPNFLRNLQEMNCDLITQAQVKAVKTHMKKSKKLDTMQQISKAGYGLLKFVTAVLGYCAVYKEVKPKKEKVEALEKEYSEAVNYLASLNREIDRLQKTLDGLNNRYETAMMRRQELQEETDLMMRRLVAADKLMSGLSSEQKRWTEDLAALYVEQSRLIGNCLLATSFLSYTGPFSFSFRQTMIYEDWLGDVMERGIPLTLPFTIERNLTNEVEVSGWNSEGLPPDELSVQNGILTTRASRFPLCIDPQTQALTWIKKKEAKNNLKVLSFNDPQFLRQLEMAIKYGMPVLFQDVNEYIDPVVDNVLEKNIKVESGRTFVMLGSTEVDYDPNFRMYLTTKLANPQFNPAAYAKAVVINYTVTVQGLEDQLLSVVVRAERSDLEEQRESLIIETSANKSLLSGLEDSLLRELATSTGNMLDNVELVNTLENTKSKAAEVRKY
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
A0A3S2LZK1
H9IX76
A0A2W1BD47
A0A2H1VR18
A0A194QEV0
A0A212EHR2
+ More
A0A2A4JUQ2 D6WU89 A0A1B6E9V8 N6TR49 U4U1Q7 B0W463 E0VQP2 A0A182NRZ8 A0A182IR22 A0A182ML65 A0A182PGJ2 A0A084WBS3 A0A2K6VB31 A0A182WSM3 A0A182TQL6 W5JGD3 A0A182HLH3 A0A182RZ92 A0A182Y3M9 Q7PUK6 A0A182QCK4 A0A182VZG6 K7J323 A0A182JSY3 A0A2R2MKY4 A0A182FQC7 A0A2T7PHE3 A0A2A3ERY9 W5MFQ0 A0A310SST6 A0A088ANX7 W4ZLH9 A0A3S1HDS1 A0A2C9JWZ8 A0A2C9JWZ0 A0A1S3PFH5 A0A1S3PEP0 A0A0N0U4V3 A0A3B4UD57 A0A0J7L4X0 V4BRT3 A0A2D0PL14 A0A3P8RA28 A0A2D0PNR9 A0A3B4YPF3 A0A3B4E236 A0A2J7QWX6 A0A0L8GLR3 A0A182VNE8 A0A1I8GN74 A0A267GW52 A0A0L8GLN5 A0A3P9CLL7 A0A1I8HCA9 A0A3B4GFI9 A0A3Q2WFN5 A0A267GS85 A0A060W723 A0A3P9A2D6 A0A1I8H2U4 A0A3Q4MJC5 A0A1L8I0W3 A0A1I8PAD1 A0A3Q0K813 E2AN69 A0A3Q0K9X2 F7DW33 A0A3Q3MNV8 A0A1A9XJK5 A0A1W4ZUV9 W5K6M6 A0A151WV17 A0A210Q0K8 A0A154PNN1 A0A1B0C738 A0A3L8DJJ8 A0A3B3TNK4 E9IZ04 A0A1I8NF00 K1R5R4 A0A452I0W3 I3K6T1 A0A452I0T0 A0A195E3E9 A0A3Q1B010 A0A1B0G3I6 A0A087XTH1 A0A1S8WG81 C3XV30 A0A3B3QK43 A0A3N0XJC6 G1NVC6 A0A1A9ZFQ2
A0A2A4JUQ2 D6WU89 A0A1B6E9V8 N6TR49 U4U1Q7 B0W463 E0VQP2 A0A182NRZ8 A0A182IR22 A0A182ML65 A0A182PGJ2 A0A084WBS3 A0A2K6VB31 A0A182WSM3 A0A182TQL6 W5JGD3 A0A182HLH3 A0A182RZ92 A0A182Y3M9 Q7PUK6 A0A182QCK4 A0A182VZG6 K7J323 A0A182JSY3 A0A2R2MKY4 A0A182FQC7 A0A2T7PHE3 A0A2A3ERY9 W5MFQ0 A0A310SST6 A0A088ANX7 W4ZLH9 A0A3S1HDS1 A0A2C9JWZ8 A0A2C9JWZ0 A0A1S3PFH5 A0A1S3PEP0 A0A0N0U4V3 A0A3B4UD57 A0A0J7L4X0 V4BRT3 A0A2D0PL14 A0A3P8RA28 A0A2D0PNR9 A0A3B4YPF3 A0A3B4E236 A0A2J7QWX6 A0A0L8GLR3 A0A182VNE8 A0A1I8GN74 A0A267GW52 A0A0L8GLN5 A0A3P9CLL7 A0A1I8HCA9 A0A3B4GFI9 A0A3Q2WFN5 A0A267GS85 A0A060W723 A0A3P9A2D6 A0A1I8H2U4 A0A3Q4MJC5 A0A1L8I0W3 A0A1I8PAD1 A0A3Q0K813 E2AN69 A0A3Q0K9X2 F7DW33 A0A3Q3MNV8 A0A1A9XJK5 A0A1W4ZUV9 W5K6M6 A0A151WV17 A0A210Q0K8 A0A154PNN1 A0A1B0C738 A0A3L8DJJ8 A0A3B3TNK4 E9IZ04 A0A1I8NF00 K1R5R4 A0A452I0W3 I3K6T1 A0A452I0T0 A0A195E3E9 A0A3Q1B010 A0A1B0G3I6 A0A087XTH1 A0A1S8WG81 C3XV30 A0A3B3QK43 A0A3N0XJC6 G1NVC6 A0A1A9ZFQ2
Pubmed
19121390
28756777
26354079
22118469
18362917
19820115
+ More
23537049 20566863 24438588 20920257 25244985 12364791 14747013 17210077 20075255 26383154 15562597 23254933 26392545 25186727 24755649 25069045 27762356 20798317 20431018 25329095 28812685 30249741 21282665 25315136 22992520 28562605 18563158 29240929 21993624
23537049 20566863 24438588 20920257 25244985 12364791 14747013 17210077 20075255 26383154 15562597 23254933 26392545 25186727 24755649 25069045 27762356 20798317 20431018 25329095 28812685 30249741 21282665 25315136 22992520 28562605 18563158 29240929 21993624
EMBL
RSAL01000096
RVE47735.1
BABH01018149
BABH01018150
BABH01018151
BABH01018152
+ More
BABH01018153 BABH01018154 BABH01018155 BABH01018156 BABH01018157 BABH01018158 KZ150261 PZC71734.1 ODYU01003927 SOQ43291.1 KQ459053 KPJ04007.1 AGBW02014810 OWR41026.1 NWSH01000608 PCG75354.1 KQ971352 EFA06783.2 GEDC01002595 JAS34703.1 APGK01057806 KB741282 ENN70796.1 KB631802 ERL86248.1 DS231835 EDS32907.1 DS235430 EEB15698.1 AXCM01008242 ATLV01022432 ATLV01022433 KE525332 KFB47667.1 ADMH02001601 ETN61879.1 APCN01000900 AAAB01008987 EAA01367.6 AXCN02000261 PZQS01000004 PVD32839.1 KZ288193 PBC34042.1 AHAT01005142 AHAT01005143 KQ759770 OAD62940.1 AAGJ04108843 AAGJ04108844 RQTK01000590 RUS77325.1 KQ435796 KOX73524.1 LBMM01000749 KMQ97578.1 KB202237 ESO91629.1 NEVH01009413 PNF33091.1 KQ421273 KOF77888.1 NIVC01000119 PAA90251.1 KOF77887.1 NIVC01000173 PAA88873.1 FR904417 CDQ62791.1 CM004466 OCU02010.1 GL441102 EFN65120.1 AAMC01042923 AAMC01042924 KQ982718 KYQ51703.1 NEDP02005302 OWF42267.1 KQ435007 KZC13452.1 JXJN01027505 JXJN01027506 JXJN01027507 QOIP01000007 RLU20605.1 GL767078 EFZ14199.1 JH817887 EKC41048.1 AERX01026833 AERX01026834 KQ979701 KYN19673.1 CCAG010016326 CCAG010016327 AYCK01007726 KV907382 OON13472.1 GG666468 EEN67956.1 RJVU01073043 ROI27764.1 AAPE02053552 AAPE02053553 AAPE02053554
BABH01018153 BABH01018154 BABH01018155 BABH01018156 BABH01018157 BABH01018158 KZ150261 PZC71734.1 ODYU01003927 SOQ43291.1 KQ459053 KPJ04007.1 AGBW02014810 OWR41026.1 NWSH01000608 PCG75354.1 KQ971352 EFA06783.2 GEDC01002595 JAS34703.1 APGK01057806 KB741282 ENN70796.1 KB631802 ERL86248.1 DS231835 EDS32907.1 DS235430 EEB15698.1 AXCM01008242 ATLV01022432 ATLV01022433 KE525332 KFB47667.1 ADMH02001601 ETN61879.1 APCN01000900 AAAB01008987 EAA01367.6 AXCN02000261 PZQS01000004 PVD32839.1 KZ288193 PBC34042.1 AHAT01005142 AHAT01005143 KQ759770 OAD62940.1 AAGJ04108843 AAGJ04108844 RQTK01000590 RUS77325.1 KQ435796 KOX73524.1 LBMM01000749 KMQ97578.1 KB202237 ESO91629.1 NEVH01009413 PNF33091.1 KQ421273 KOF77888.1 NIVC01000119 PAA90251.1 KOF77887.1 NIVC01000173 PAA88873.1 FR904417 CDQ62791.1 CM004466 OCU02010.1 GL441102 EFN65120.1 AAMC01042923 AAMC01042924 KQ982718 KYQ51703.1 NEDP02005302 OWF42267.1 KQ435007 KZC13452.1 JXJN01027505 JXJN01027506 JXJN01027507 QOIP01000007 RLU20605.1 GL767078 EFZ14199.1 JH817887 EKC41048.1 AERX01026833 AERX01026834 KQ979701 KYN19673.1 CCAG010016326 CCAG010016327 AYCK01007726 KV907382 OON13472.1 GG666468 EEN67956.1 RJVU01073043 ROI27764.1 AAPE02053552 AAPE02053553 AAPE02053554
Proteomes
UP000283053
UP000005204
UP000053268
UP000007151
UP000218220
UP000007266
+ More
UP000019118 UP000030742 UP000002320 UP000009046 UP000075884 UP000075880 UP000075883 UP000075885 UP000030765 UP000076407 UP000075902 UP000000673 UP000075840 UP000075900 UP000076408 UP000007062 UP000075886 UP000075920 UP000002358 UP000075881 UP000085678 UP000069272 UP000245119 UP000242457 UP000018468 UP000005203 UP000007110 UP000271974 UP000076420 UP000087266 UP000053105 UP000261420 UP000036403 UP000030746 UP000221080 UP000265100 UP000261360 UP000261440 UP000235965 UP000053454 UP000075903 UP000095280 UP000215902 UP000265160 UP000261460 UP000264840 UP000193380 UP000265140 UP000261580 UP000186698 UP000095300 UP000000311 UP000008143 UP000261640 UP000092443 UP000192224 UP000018467 UP000075809 UP000242188 UP000076502 UP000092460 UP000279307 UP000261500 UP000095301 UP000005408 UP000291020 UP000005207 UP000078492 UP000257160 UP000092444 UP000028760 UP000001554 UP000261540 UP000001074 UP000092445
UP000019118 UP000030742 UP000002320 UP000009046 UP000075884 UP000075880 UP000075883 UP000075885 UP000030765 UP000076407 UP000075902 UP000000673 UP000075840 UP000075900 UP000076408 UP000007062 UP000075886 UP000075920 UP000002358 UP000075881 UP000085678 UP000069272 UP000245119 UP000242457 UP000018468 UP000005203 UP000007110 UP000271974 UP000076420 UP000087266 UP000053105 UP000261420 UP000036403 UP000030746 UP000221080 UP000265100 UP000261360 UP000261440 UP000235965 UP000053454 UP000075903 UP000095280 UP000215902 UP000265160 UP000261460 UP000264840 UP000193380 UP000265140 UP000261580 UP000186698 UP000095300 UP000000311 UP000008143 UP000261640 UP000092443 UP000192224 UP000018467 UP000075809 UP000242188 UP000076502 UP000092460 UP000279307 UP000261500 UP000095301 UP000005408 UP000291020 UP000005207 UP000078492 UP000257160 UP000092444 UP000028760 UP000001554 UP000261540 UP000001074 UP000092445
Pfam
Interpro
IPR035699
AAA_6
+ More
IPR042228 Dynein_2_C
IPR041228 Dynein_C
IPR041658 AAA_lid_11
IPR003593 AAA+_ATPase
IPR041466 Dynein_AAA5_ext
IPR013594 Dynein_heavy_dom-1
IPR004273 Dynein_heavy_D6_P-loop
IPR013602 Dynein_heavy_dom-2
IPR042219 AAA_lid_11_sf
IPR024743 Dynein_HC_stalk
IPR024317 Dynein_heavy_chain_D4_dom
IPR027417 P-loop_NTPase
IPR042222 Dynein_2_N
IPR041589 DNAH3_AAA_lid_1
IPR035706 AAA_9
IPR026983 DHC_fam
IPR011704 ATPase_dyneun-rel_AAA
IPR000276 GPCR_Rhodpsn
IPR017452 GPCR_Rhodpsn_7TM
IPR018130 Ribosomal_S2_CS
IPR002108 ADF-H
IPR017904 ADF/Cofilin
IPR017438 ATP-NAD_kinase_N
IPR016064 NAD/diacylglycerol_kinase_sf
IPR001206 Diacylglycerol_kinase_cat_dom
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR027267 AH/BAR_dom_sf
IPR042228 Dynein_2_C
IPR041228 Dynein_C
IPR041658 AAA_lid_11
IPR003593 AAA+_ATPase
IPR041466 Dynein_AAA5_ext
IPR013594 Dynein_heavy_dom-1
IPR004273 Dynein_heavy_D6_P-loop
IPR013602 Dynein_heavy_dom-2
IPR042219 AAA_lid_11_sf
IPR024743 Dynein_HC_stalk
IPR024317 Dynein_heavy_chain_D4_dom
IPR027417 P-loop_NTPase
IPR042222 Dynein_2_N
IPR041589 DNAH3_AAA_lid_1
IPR035706 AAA_9
IPR026983 DHC_fam
IPR011704 ATPase_dyneun-rel_AAA
IPR000276 GPCR_Rhodpsn
IPR017452 GPCR_Rhodpsn_7TM
IPR018130 Ribosomal_S2_CS
IPR002108 ADF-H
IPR017904 ADF/Cofilin
IPR017438 ATP-NAD_kinase_N
IPR016064 NAD/diacylglycerol_kinase_sf
IPR001206 Diacylglycerol_kinase_cat_dom
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR027267 AH/BAR_dom_sf
ProteinModelPortal
A0A3S2LZK1
H9IX76
A0A2W1BD47
A0A2H1VR18
A0A194QEV0
A0A212EHR2
+ More
A0A2A4JUQ2 D6WU89 A0A1B6E9V8 N6TR49 U4U1Q7 B0W463 E0VQP2 A0A182NRZ8 A0A182IR22 A0A182ML65 A0A182PGJ2 A0A084WBS3 A0A2K6VB31 A0A182WSM3 A0A182TQL6 W5JGD3 A0A182HLH3 A0A182RZ92 A0A182Y3M9 Q7PUK6 A0A182QCK4 A0A182VZG6 K7J323 A0A182JSY3 A0A2R2MKY4 A0A182FQC7 A0A2T7PHE3 A0A2A3ERY9 W5MFQ0 A0A310SST6 A0A088ANX7 W4ZLH9 A0A3S1HDS1 A0A2C9JWZ8 A0A2C9JWZ0 A0A1S3PFH5 A0A1S3PEP0 A0A0N0U4V3 A0A3B4UD57 A0A0J7L4X0 V4BRT3 A0A2D0PL14 A0A3P8RA28 A0A2D0PNR9 A0A3B4YPF3 A0A3B4E236 A0A2J7QWX6 A0A0L8GLR3 A0A182VNE8 A0A1I8GN74 A0A267GW52 A0A0L8GLN5 A0A3P9CLL7 A0A1I8HCA9 A0A3B4GFI9 A0A3Q2WFN5 A0A267GS85 A0A060W723 A0A3P9A2D6 A0A1I8H2U4 A0A3Q4MJC5 A0A1L8I0W3 A0A1I8PAD1 A0A3Q0K813 E2AN69 A0A3Q0K9X2 F7DW33 A0A3Q3MNV8 A0A1A9XJK5 A0A1W4ZUV9 W5K6M6 A0A151WV17 A0A210Q0K8 A0A154PNN1 A0A1B0C738 A0A3L8DJJ8 A0A3B3TNK4 E9IZ04 A0A1I8NF00 K1R5R4 A0A452I0W3 I3K6T1 A0A452I0T0 A0A195E3E9 A0A3Q1B010 A0A1B0G3I6 A0A087XTH1 A0A1S8WG81 C3XV30 A0A3B3QK43 A0A3N0XJC6 G1NVC6 A0A1A9ZFQ2
A0A2A4JUQ2 D6WU89 A0A1B6E9V8 N6TR49 U4U1Q7 B0W463 E0VQP2 A0A182NRZ8 A0A182IR22 A0A182ML65 A0A182PGJ2 A0A084WBS3 A0A2K6VB31 A0A182WSM3 A0A182TQL6 W5JGD3 A0A182HLH3 A0A182RZ92 A0A182Y3M9 Q7PUK6 A0A182QCK4 A0A182VZG6 K7J323 A0A182JSY3 A0A2R2MKY4 A0A182FQC7 A0A2T7PHE3 A0A2A3ERY9 W5MFQ0 A0A310SST6 A0A088ANX7 W4ZLH9 A0A3S1HDS1 A0A2C9JWZ8 A0A2C9JWZ0 A0A1S3PFH5 A0A1S3PEP0 A0A0N0U4V3 A0A3B4UD57 A0A0J7L4X0 V4BRT3 A0A2D0PL14 A0A3P8RA28 A0A2D0PNR9 A0A3B4YPF3 A0A3B4E236 A0A2J7QWX6 A0A0L8GLR3 A0A182VNE8 A0A1I8GN74 A0A267GW52 A0A0L8GLN5 A0A3P9CLL7 A0A1I8HCA9 A0A3B4GFI9 A0A3Q2WFN5 A0A267GS85 A0A060W723 A0A3P9A2D6 A0A1I8H2U4 A0A3Q4MJC5 A0A1L8I0W3 A0A1I8PAD1 A0A3Q0K813 E2AN69 A0A3Q0K9X2 F7DW33 A0A3Q3MNV8 A0A1A9XJK5 A0A1W4ZUV9 W5K6M6 A0A151WV17 A0A210Q0K8 A0A154PNN1 A0A1B0C738 A0A3L8DJJ8 A0A3B3TNK4 E9IZ04 A0A1I8NF00 K1R5R4 A0A452I0W3 I3K6T1 A0A452I0T0 A0A195E3E9 A0A3Q1B010 A0A1B0G3I6 A0A087XTH1 A0A1S8WG81 C3XV30 A0A3B3QK43 A0A3N0XJC6 G1NVC6 A0A1A9ZFQ2
PDB
3VKH
E-value=9.05535e-85,
Score=802
Ontologies
GO
PANTHER
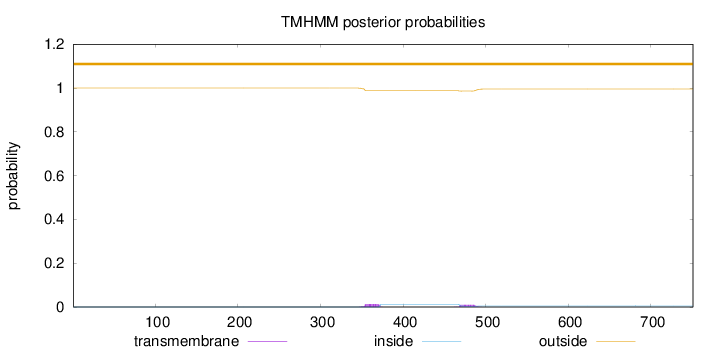
Topology
Length:
751
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.46573
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00020
outside
1 - 751
Population Genetic Test Statistics
Pi
174.166771
Theta
152.354652
Tajima's D
0.604931
CLR
1.260719
CSRT
0.542422878856057
Interpretation
Uncertain
