Pre Gene Modal
BGIBMGA001859
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_dynein_heavy_chain_10?_axonemal-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.77
Sequence
CDS
ATGCAGCTCGGAGGAGCTCCAGCCGGTCCCGCTGGTACCGGCAAAACCGAAACTACAAAAGATTTAGCAAAGGCTTTAGGTCTTCTATGCGTGGTGACCAACTGCGGGGAGGGAATGGATTTCCGAGCTGTTGGACAGATCCTCGCTGGCCTTTGCCAGTGCGGTGCTTGGGGATGCTTCGACGAATTCAACAGGATTGATATATCAGTGCTGTCAGTTATTTCAACGCAGCTACAGTGCATCAGAAGTGCTTTGCTCATGAAATTGAAACGTTTCACGTTTGAAGGGCAGGAGATAGCAATGGATAGCAAAGTTGGTATCTTTATCACCATGAATCCTGGATACGCCGGTCGAACGGAGTTACCTGAATCCGTGAAAGCTTTGTTCAGACCTGTCGTGTGTATTCTACCTGACCTTGAAATGATTTGTCAGATATCCCTATTCTCTGATGGATTTCTTACGGCCAAAGTTCTAGCCAAGAAAATGACAGTCCTATACAAAGTAGCAAGAGAACAACTATCAAAACAATCTCACTATGACTGGGGATTAAGGGCTCTAACGGCGGTCCTCAGGATGGCGGGGAAATTGAGAAGAGATTCCCCAGGACTCAGTGAAATTATGGTGCTGATGAGAGCTTTAAGGGACATGAACCATCCAAAATTCGTATTTGAAGACGTGCCTCTATTTCTTGGACTGATAAAAGACCTTTTCCCTGGGTTGGAGTGTCCTCGTGTTGGTTATCCAGAATTCAATGCTGCCGTTCTTGAGGTTTTAGAAAAAGATGGATATGTTGTACTACCGCATCAGGTTGATAAAGTGGTTCAACTTTACGAAACGATGATGACTAGACACTGTACGATGCTAGTGGGGCCCACGGGCGGCGGCAAGACGGTTATACTTCATTGTCTAGTGAAAGCTCAAACTAATTTAGGTCTACCTACAAAACTTACAGTTGTTAATCCAAAGGCCTGCTCCGTTATAGAGCTGTATGGTATCTTGGACCCTGTAACAAGAGACTGGACCGATGGTTTATATTCAAAAATATTTAGGGAAATGAACAGACCGGCAGAAAAGAATGAGCGTCGCTACTCATTATTCGATGGAGATGTAGACGCTCTTTGGATAGAGAACATGAATTCGGTAATGGACGATAACAAGCTCTTGACCCTCGCGAACGGAGAGCGAATCAGACTGGCCCCGTACTGCTCTTTACTCTTCGAAGTTGGAGATTTGAACTATGCCTCTCCGGCCACTGTTTCCAGAGCCGGAATGGTGTTCGTCGACCCTAAGAACTTGGGATACGAACCTTACTGGGAAAGGCACTTGTAA
Protein
MQLGGAPAGPAGTGKTETTKDLAKALGLLCVVTNCGEGMDFRAVGQILAGLCQCGAWGCFDEFNRIDISVLSVISTQLQCIRSALLMKLKRFTFEGQEIAMDSKVGIFITMNPGYAGRTELPESVKALFRPVVCILPDLEMICQISLFSDGFLTAKVLAKKMTVLYKVAREQLSKQSHYDWGLRALTAVLRMAGKLRRDSPGLSEIMVLMRALRDMNHPKFVFEDVPLFLGLIKDLFPGLECPRVGYPEFNAAVLEVLEKDGYVVLPHQVDKVVQLYETMMTRHCTMLVGPTGGGKTVILHCLVKAQTNLGLPTKLTVVNPKACSVIELYGILDPVTRDWTDGLYSKIFREMNRPAEKNERRYSLFDGDVDALWIENMNSVMDDNKLLTLANGERIRLAPYCSLLFEVGDLNYASPATVSRAGMVFVDPKNLGYEPYWERHL
Summary
Uniprot
H9IX76
A0A2A4J3H3
A0A212EHR2
A0A194RBR8
A0A194QEV0
A0A2H1VR18
+ More
A0A2W1BD47 A0A3S2LZK1 U4U1Q7 N6TR49 D6WU89 A0A1A9UNN9 A0A1W4W2L4 E0VQP2 B4MC48 E1JJ04 Q9VAV5 B4IH10 A0A1W4WEG6 A0A3B0JD27 B4PQS1 B4N8N4 A0A182IR22 B4JGP4 A0A0B4KI48 A0A1B0C4S2 A0A0M4EPR1 Q7PUK6 A0A182L9W2 A0A182VZG6 A0A182RZ92 A0A182SS90 A0A182NRZ8 B4G3H4 B4K6Z4 I5ANL7 A0A1B6E9V8 A0A0Q9X9I4 B3LV78 A0A182PGJ2 A0A1B0FPJ7 A0A1B6DAB9 A0A1B6FUW5 A0A182TQL6 Q299Y5 A0A182QCK4 A0A1I8PAD1 A0A3M0JXR0 A0A182JSY3 A0A2C9JWZ8 A0A2C9JWZ0 A0A1D5PD16 B0W463 A0A1V4JSE1 A0A084WBS4 R7UWA6 A0A3L8S573 A0A151NS45 E7FGT8 A0A218UMB0 F7DW33 A0A1I8GN74 A0A267GW52 I3K6T1 A0A452I0T0 A0A452I0W3 W5K6M6 A0A1U8CX33 A0A091FLU3 A0A3Q2WFN5 R0LTG7 U3J391 U3K3Y5 A0A2I0M866 A0A087XTH1 A0A2G9SBI3 K1R5R4 A0A091VYL6 A0A3B3TNK4 A0A1J1J5S5 A0A1I8HCA9 Q27806 A0A3Q3MNV8 A0A094K0P6 A0A3B4E236 A0A3P9CLL7 G3P1E8 A0A3Q4MJC5 A0A287AF97 F1RFM8 A0A3B4GFI9 A0A2Y9FL08 A0A336KTI1 W5MFQ0 A0A3B4YPF3 H9GM01 A0A2A3ERY9 C3XV30 A0A1S3W223 A0A384AQQ9
A0A2W1BD47 A0A3S2LZK1 U4U1Q7 N6TR49 D6WU89 A0A1A9UNN9 A0A1W4W2L4 E0VQP2 B4MC48 E1JJ04 Q9VAV5 B4IH10 A0A1W4WEG6 A0A3B0JD27 B4PQS1 B4N8N4 A0A182IR22 B4JGP4 A0A0B4KI48 A0A1B0C4S2 A0A0M4EPR1 Q7PUK6 A0A182L9W2 A0A182VZG6 A0A182RZ92 A0A182SS90 A0A182NRZ8 B4G3H4 B4K6Z4 I5ANL7 A0A1B6E9V8 A0A0Q9X9I4 B3LV78 A0A182PGJ2 A0A1B0FPJ7 A0A1B6DAB9 A0A1B6FUW5 A0A182TQL6 Q299Y5 A0A182QCK4 A0A1I8PAD1 A0A3M0JXR0 A0A182JSY3 A0A2C9JWZ8 A0A2C9JWZ0 A0A1D5PD16 B0W463 A0A1V4JSE1 A0A084WBS4 R7UWA6 A0A3L8S573 A0A151NS45 E7FGT8 A0A218UMB0 F7DW33 A0A1I8GN74 A0A267GW52 I3K6T1 A0A452I0T0 A0A452I0W3 W5K6M6 A0A1U8CX33 A0A091FLU3 A0A3Q2WFN5 R0LTG7 U3J391 U3K3Y5 A0A2I0M866 A0A087XTH1 A0A2G9SBI3 K1R5R4 A0A091VYL6 A0A3B3TNK4 A0A1J1J5S5 A0A1I8HCA9 Q27806 A0A3Q3MNV8 A0A094K0P6 A0A3B4E236 A0A3P9CLL7 G3P1E8 A0A3Q4MJC5 A0A287AF97 F1RFM8 A0A3B4GFI9 A0A2Y9FL08 A0A336KTI1 W5MFQ0 A0A3B4YPF3 H9GM01 A0A2A3ERY9 C3XV30 A0A1S3W223 A0A384AQQ9
Pubmed
19121390
22118469
26354079
28756777
23537049
18362917
+ More
19820115 20566863 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 12364791 14747013 17210077 20966253 15632085 23185243 15562597 15592404 24438588 23254933 30282656 22293439 23594743 20431018 26392545 25186727 28562605 25329095 23749191 23371554 22992520 8186465 21881562 18563158
19820115 20566863 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 12364791 14747013 17210077 20966253 15632085 23185243 15562597 15592404 24438588 23254933 30282656 22293439 23594743 20431018 26392545 25186727 28562605 25329095 23749191 23371554 22992520 8186465 21881562 18563158
EMBL
BABH01018149
BABH01018150
BABH01018151
BABH01018152
BABH01018153
BABH01018154
+ More
BABH01018155 BABH01018156 BABH01018157 BABH01018158 NWSH01003463 PCG66238.1 AGBW02014810 OWR41026.1 KQ460436 KPJ14720.1 KQ459053 KPJ04007.1 ODYU01003927 SOQ43291.1 KZ150261 PZC71734.1 RSAL01000096 RVE47735.1 KB631802 ERL86248.1 APGK01057806 KB741282 ENN70796.1 KQ971352 EFA06783.2 DS235430 EEB15698.1 CH940656 EDW58669.2 AE014297 ACZ95053.1 AAF56793.3 CH480837 EDW49128.1 OUUW01000005 SPP80284.1 CM000160 EDW98410.2 CH964232 EDW81485.2 CH916369 EDV92648.1 AGB96416.1 JXJN01025625 JXJN01025626 JXJN01025627 CP012526 ALC46147.1 AAAB01008987 EAA01367.6 CH479179 EDW24982.1 CH933806 EDW15281.2 CM000070 EIM52552.2 GEDC01002595 JAS34703.1 KRG01490.1 CH902617 EDV43610.2 CCAG010022515 GEDC01014670 JAS22628.1 GECZ01015781 JAS53988.1 EAL27566.4 EIM52553.2 AXCN02000261 QRBI01000121 RMC05733.1 AADN05000505 DS231835 EDS32907.1 LSYS01006629 OPJ75091.1 ATLV01022433 ATLV01022434 KE525332 KFB47668.1 AMQN01006005 KB297391 ELU10552.1 QUSF01000066 RLV96891.1 AKHW03002185 KYO39614.1 BX323557 MUZQ01000225 OWK54873.1 AAMC01042923 AAMC01042924 NIVC01000119 PAA90251.1 AERX01026833 AERX01026834 KL447264 KFO71450.1 KB742840 EOB03708.1 ADON01049977 ADON01049978 ADON01049979 AGTO01010316 AKCR02000030 PKK25878.1 AYCK01007726 KV926115 PIO37502.1 JH817887 EKC41048.1 KK734463 KFR08294.1 CVRI01000070 CRL07346.1 U03973 AAA63587.1 KL336764 KFZ50352.1 AEMK02000096 UFQS01000469 UFQT01000469 SSX04260.1 SSX24625.1 AHAT01005142 AHAT01005143 KZ288193 PBC34042.1 GG666468 EEN67956.1
BABH01018155 BABH01018156 BABH01018157 BABH01018158 NWSH01003463 PCG66238.1 AGBW02014810 OWR41026.1 KQ460436 KPJ14720.1 KQ459053 KPJ04007.1 ODYU01003927 SOQ43291.1 KZ150261 PZC71734.1 RSAL01000096 RVE47735.1 KB631802 ERL86248.1 APGK01057806 KB741282 ENN70796.1 KQ971352 EFA06783.2 DS235430 EEB15698.1 CH940656 EDW58669.2 AE014297 ACZ95053.1 AAF56793.3 CH480837 EDW49128.1 OUUW01000005 SPP80284.1 CM000160 EDW98410.2 CH964232 EDW81485.2 CH916369 EDV92648.1 AGB96416.1 JXJN01025625 JXJN01025626 JXJN01025627 CP012526 ALC46147.1 AAAB01008987 EAA01367.6 CH479179 EDW24982.1 CH933806 EDW15281.2 CM000070 EIM52552.2 GEDC01002595 JAS34703.1 KRG01490.1 CH902617 EDV43610.2 CCAG010022515 GEDC01014670 JAS22628.1 GECZ01015781 JAS53988.1 EAL27566.4 EIM52553.2 AXCN02000261 QRBI01000121 RMC05733.1 AADN05000505 DS231835 EDS32907.1 LSYS01006629 OPJ75091.1 ATLV01022433 ATLV01022434 KE525332 KFB47668.1 AMQN01006005 KB297391 ELU10552.1 QUSF01000066 RLV96891.1 AKHW03002185 KYO39614.1 BX323557 MUZQ01000225 OWK54873.1 AAMC01042923 AAMC01042924 NIVC01000119 PAA90251.1 AERX01026833 AERX01026834 KL447264 KFO71450.1 KB742840 EOB03708.1 ADON01049977 ADON01049978 ADON01049979 AGTO01010316 AKCR02000030 PKK25878.1 AYCK01007726 KV926115 PIO37502.1 JH817887 EKC41048.1 KK734463 KFR08294.1 CVRI01000070 CRL07346.1 U03973 AAA63587.1 KL336764 KFZ50352.1 AEMK02000096 UFQS01000469 UFQT01000469 SSX04260.1 SSX24625.1 AHAT01005142 AHAT01005143 KZ288193 PBC34042.1 GG666468 EEN67956.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000030742 UP000019118 UP000007266 UP000078200 UP000192221 UP000009046 UP000008792 UP000000803 UP000001292 UP000268350 UP000002282 UP000007798 UP000075880 UP000001070 UP000092460 UP000092553 UP000007062 UP000075882 UP000075920 UP000075900 UP000075901 UP000075884 UP000008744 UP000009192 UP000001819 UP000007801 UP000075885 UP000092444 UP000075902 UP000075886 UP000095300 UP000269221 UP000075881 UP000076420 UP000000539 UP000002320 UP000190648 UP000030765 UP000014760 UP000276834 UP000050525 UP000000437 UP000197619 UP000008143 UP000095280 UP000215902 UP000005207 UP000291020 UP000018467 UP000189705 UP000053760 UP000264840 UP000016666 UP000016665 UP000053872 UP000028760 UP000005408 UP000053605 UP000261500 UP000183832 UP000261640 UP000261440 UP000265160 UP000007635 UP000261580 UP000008227 UP000261460 UP000018468 UP000261360 UP000001646 UP000242457 UP000001554 UP000079721 UP000261681
UP000030742 UP000019118 UP000007266 UP000078200 UP000192221 UP000009046 UP000008792 UP000000803 UP000001292 UP000268350 UP000002282 UP000007798 UP000075880 UP000001070 UP000092460 UP000092553 UP000007062 UP000075882 UP000075920 UP000075900 UP000075901 UP000075884 UP000008744 UP000009192 UP000001819 UP000007801 UP000075885 UP000092444 UP000075902 UP000075886 UP000095300 UP000269221 UP000075881 UP000076420 UP000000539 UP000002320 UP000190648 UP000030765 UP000014760 UP000276834 UP000050525 UP000000437 UP000197619 UP000008143 UP000095280 UP000215902 UP000005207 UP000291020 UP000018467 UP000189705 UP000053760 UP000264840 UP000016666 UP000016665 UP000053872 UP000028760 UP000005408 UP000053605 UP000261500 UP000183832 UP000261640 UP000261440 UP000265160 UP000007635 UP000261580 UP000008227 UP000261460 UP000018468 UP000261360 UP000001646 UP000242457 UP000001554 UP000079721 UP000261681
Pfam
Interpro
IPR042222
Dynein_2_N
+ More
IPR041589 DNAH3_AAA_lid_1
IPR035706 AAA_9
IPR024317 Dynein_heavy_chain_D4_dom
IPR024743 Dynein_HC_stalk
IPR027417 P-loop_NTPase
IPR041466 Dynein_AAA5_ext
IPR003593 AAA+_ATPase
IPR013594 Dynein_heavy_dom-1
IPR042219 AAA_lid_11_sf
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR035699 AAA_6
IPR041228 Dynein_C
IPR042228 Dynein_2_C
IPR041658 AAA_lid_11
IPR026983 DHC_fam
IPR002108 ADF-H
IPR017904 ADF/Cofilin
IPR017438 ATP-NAD_kinase_N
IPR016064 NAD/diacylglycerol_kinase_sf
IPR001206 Diacylglycerol_kinase_cat_dom
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR041589 DNAH3_AAA_lid_1
IPR035706 AAA_9
IPR024317 Dynein_heavy_chain_D4_dom
IPR024743 Dynein_HC_stalk
IPR027417 P-loop_NTPase
IPR041466 Dynein_AAA5_ext
IPR003593 AAA+_ATPase
IPR013594 Dynein_heavy_dom-1
IPR042219 AAA_lid_11_sf
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR035699 AAA_6
IPR041228 Dynein_C
IPR042228 Dynein_2_C
IPR041658 AAA_lid_11
IPR026983 DHC_fam
IPR002108 ADF-H
IPR017904 ADF/Cofilin
IPR017438 ATP-NAD_kinase_N
IPR016064 NAD/diacylglycerol_kinase_sf
IPR001206 Diacylglycerol_kinase_cat_dom
IPR029006 ADF-H/Gelsolin-like_dom_sf
ProteinModelPortal
H9IX76
A0A2A4J3H3
A0A212EHR2
A0A194RBR8
A0A194QEV0
A0A2H1VR18
+ More
A0A2W1BD47 A0A3S2LZK1 U4U1Q7 N6TR49 D6WU89 A0A1A9UNN9 A0A1W4W2L4 E0VQP2 B4MC48 E1JJ04 Q9VAV5 B4IH10 A0A1W4WEG6 A0A3B0JD27 B4PQS1 B4N8N4 A0A182IR22 B4JGP4 A0A0B4KI48 A0A1B0C4S2 A0A0M4EPR1 Q7PUK6 A0A182L9W2 A0A182VZG6 A0A182RZ92 A0A182SS90 A0A182NRZ8 B4G3H4 B4K6Z4 I5ANL7 A0A1B6E9V8 A0A0Q9X9I4 B3LV78 A0A182PGJ2 A0A1B0FPJ7 A0A1B6DAB9 A0A1B6FUW5 A0A182TQL6 Q299Y5 A0A182QCK4 A0A1I8PAD1 A0A3M0JXR0 A0A182JSY3 A0A2C9JWZ8 A0A2C9JWZ0 A0A1D5PD16 B0W463 A0A1V4JSE1 A0A084WBS4 R7UWA6 A0A3L8S573 A0A151NS45 E7FGT8 A0A218UMB0 F7DW33 A0A1I8GN74 A0A267GW52 I3K6T1 A0A452I0T0 A0A452I0W3 W5K6M6 A0A1U8CX33 A0A091FLU3 A0A3Q2WFN5 R0LTG7 U3J391 U3K3Y5 A0A2I0M866 A0A087XTH1 A0A2G9SBI3 K1R5R4 A0A091VYL6 A0A3B3TNK4 A0A1J1J5S5 A0A1I8HCA9 Q27806 A0A3Q3MNV8 A0A094K0P6 A0A3B4E236 A0A3P9CLL7 G3P1E8 A0A3Q4MJC5 A0A287AF97 F1RFM8 A0A3B4GFI9 A0A2Y9FL08 A0A336KTI1 W5MFQ0 A0A3B4YPF3 H9GM01 A0A2A3ERY9 C3XV30 A0A1S3W223 A0A384AQQ9
A0A2W1BD47 A0A3S2LZK1 U4U1Q7 N6TR49 D6WU89 A0A1A9UNN9 A0A1W4W2L4 E0VQP2 B4MC48 E1JJ04 Q9VAV5 B4IH10 A0A1W4WEG6 A0A3B0JD27 B4PQS1 B4N8N4 A0A182IR22 B4JGP4 A0A0B4KI48 A0A1B0C4S2 A0A0M4EPR1 Q7PUK6 A0A182L9W2 A0A182VZG6 A0A182RZ92 A0A182SS90 A0A182NRZ8 B4G3H4 B4K6Z4 I5ANL7 A0A1B6E9V8 A0A0Q9X9I4 B3LV78 A0A182PGJ2 A0A1B0FPJ7 A0A1B6DAB9 A0A1B6FUW5 A0A182TQL6 Q299Y5 A0A182QCK4 A0A1I8PAD1 A0A3M0JXR0 A0A182JSY3 A0A2C9JWZ8 A0A2C9JWZ0 A0A1D5PD16 B0W463 A0A1V4JSE1 A0A084WBS4 R7UWA6 A0A3L8S573 A0A151NS45 E7FGT8 A0A218UMB0 F7DW33 A0A1I8GN74 A0A267GW52 I3K6T1 A0A452I0T0 A0A452I0W3 W5K6M6 A0A1U8CX33 A0A091FLU3 A0A3Q2WFN5 R0LTG7 U3J391 U3K3Y5 A0A2I0M866 A0A087XTH1 A0A2G9SBI3 K1R5R4 A0A091VYL6 A0A3B3TNK4 A0A1J1J5S5 A0A1I8HCA9 Q27806 A0A3Q3MNV8 A0A094K0P6 A0A3B4E236 A0A3P9CLL7 G3P1E8 A0A3Q4MJC5 A0A287AF97 F1RFM8 A0A3B4GFI9 A0A2Y9FL08 A0A336KTI1 W5MFQ0 A0A3B4YPF3 H9GM01 A0A2A3ERY9 C3XV30 A0A1S3W223 A0A384AQQ9
PDB
4RH7
E-value=3.73151e-100,
Score=932
Ontologies
GO
PANTHER
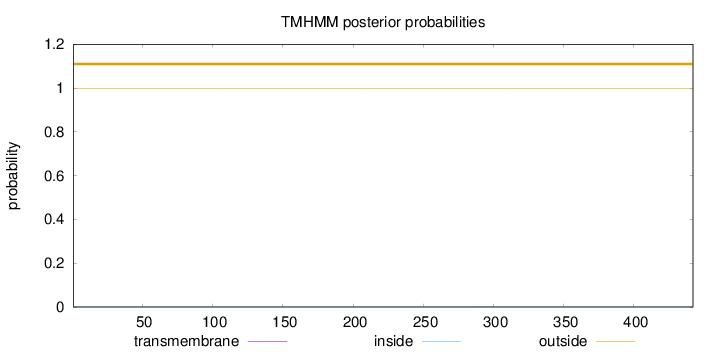
Topology
Length:
442
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01154
Exp number, first 60 AAs:
0.00348
Total prob of N-in:
0.00078
outside
1 - 442
Population Genetic Test Statistics
Pi
273.316617
Theta
171.259321
Tajima's D
1.586384
CLR
0.644548
CSRT
0.805109744512774
Interpretation
Uncertain
